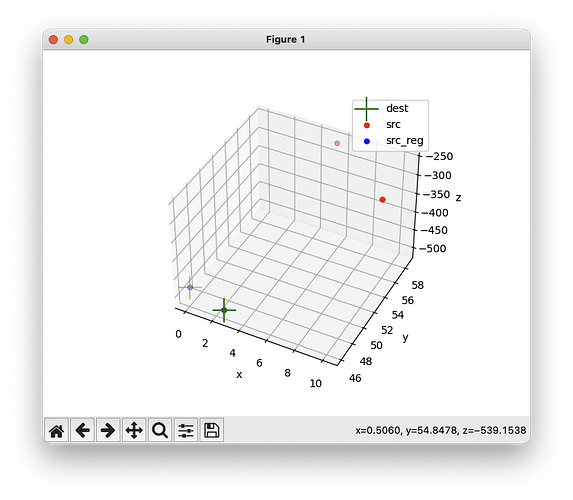
Hi @kuech,
Again, very sorry about the delayed response-- here are some answers:
Interestingly it doesn’t work when I add the labels to the PAM50_label_body.nii.gz file and use the -l argument during registration.
Indeed, this is strange. We have opened an issue about it and we will investigate: ERROR: Wrong landmarks input. Labels must have correspondence in template space. · Issue #4227 · spinalcordtoolbox/spinalcordtoolbox · GitHub
Placing a single voxel on that cord mask-slice was not sufficient in some subjects. It still got lost during straightening
As mentioned above (Registration of lumbar data to PAM50 - #2 by jcohenadad), I strongly recommend not involving straightening in the registration process, precisely because of the issues you are observing (among other reasons).
Yes, 0.5x0.5x5mm. I tried this on a few subjects now, but it produces something wrong or gets stuck. I let it run for 20min before interrupting. Error happens during estimate transformation for step #2.
In one case the registration rotates the image by about 45° degrees. I added a folder containing this example with the code for reproduction (including the new label files). I guess the cost function gets stuck in a local minima or trapped without reaching exit-conditions? In which case the registration to the template seems to be more robust or even the only option. example_wcode.zip (804.1 KB)
Thank you for providing the data for reproducing. Trying the commands in your script, with comments below:
 I recommend using the
I recommend using the -qc flag, especially when optimizing parameters. You can quickly have an overview of the result, while keeping track of the syntax for each combination of parameters.
-ref template
sct_register_to_template -i $anat -s $mask -ldisc $label -c t2 -ref template -ofolder reg_to_tem
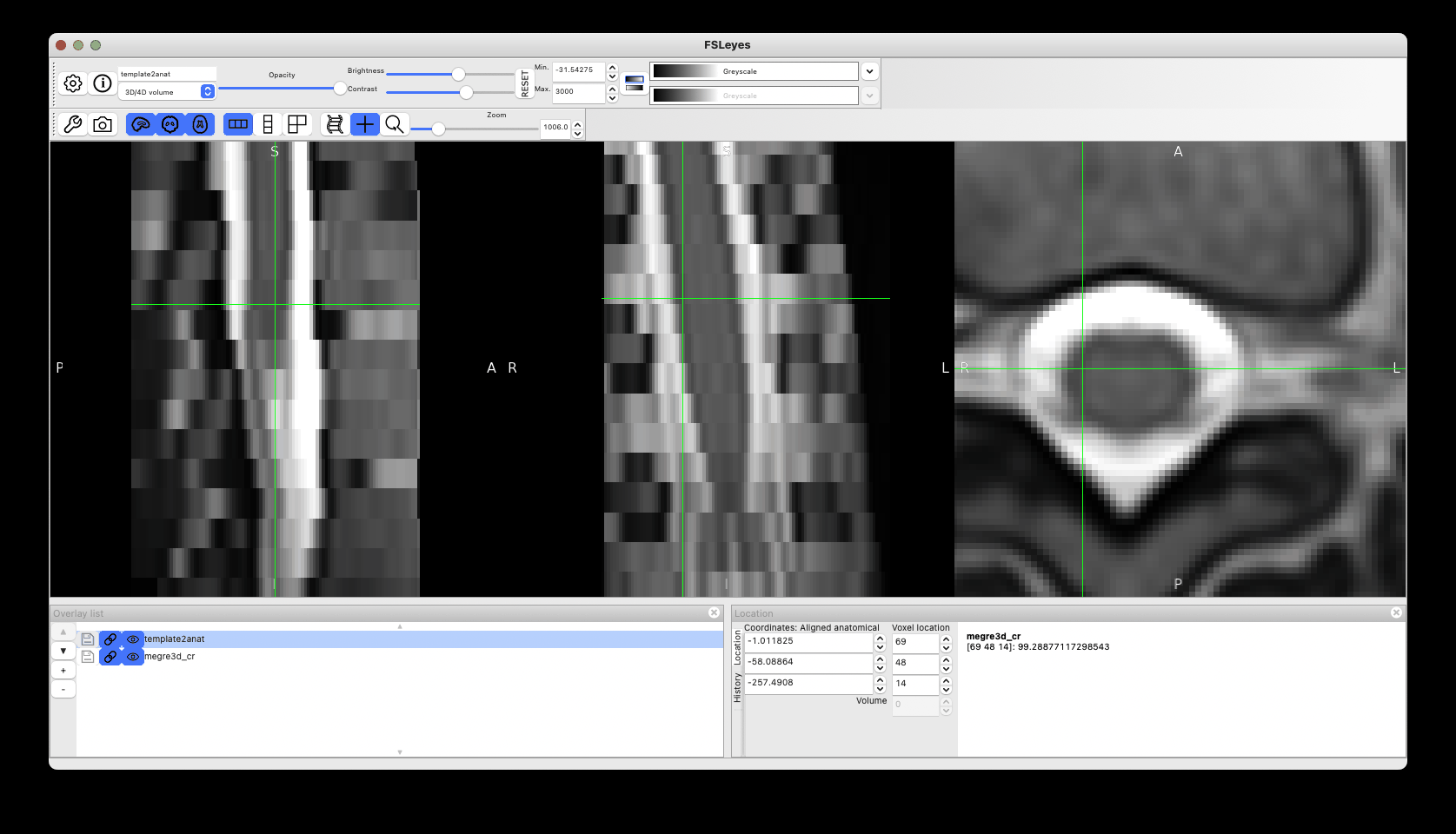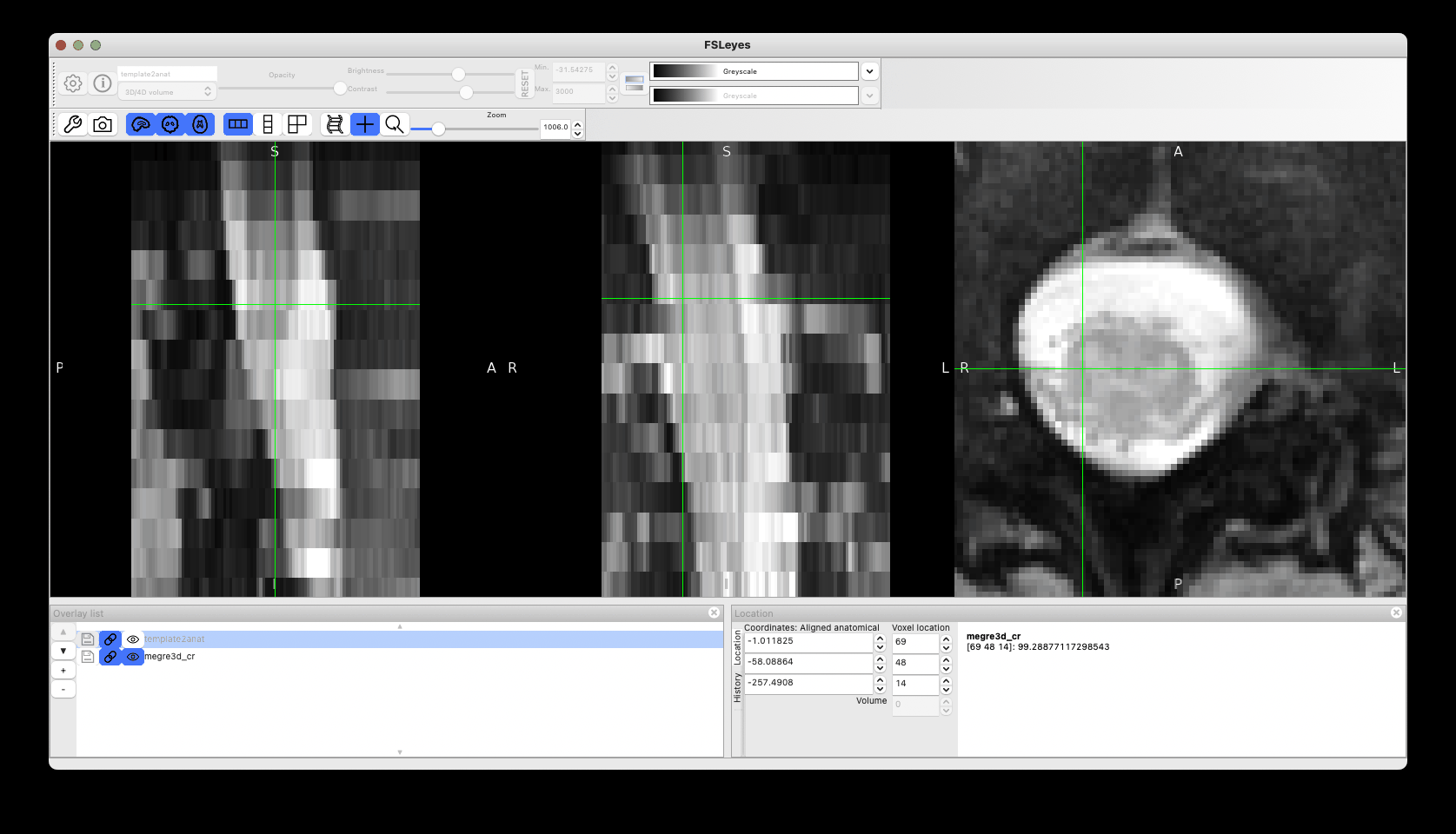
This produces reasonable registration (with room for improvements by tweaking the registration params), although as discussed above I would avoid straightening.
-ref template with flag -l
Comment from the SHELL script:
Using the -l argument: will not run!
I can reproduce, it does not run. More specifically, here is the full Terminal output:
Terminal output
julien-macbook:~/Desktop/example_wcode $ sct_register_to_template -i megre3d_cr.nii.gz -s megre3d_cr_msc.nii.gz -l megre3d_cr_label.nii.gz -c t2 -ref template -ofolder reg_to_tem_body
--
Spinal Cord Toolbox (git-master-76c27e16f13974de3ff8f81e25c92abe8f6d80ec)
sct_register_to_template -i megre3d_cr.nii.gz -s megre3d_cr_msc.nii.gz -l megre3d_cr_label.nii.gz -c t2 -ref template -ofolder reg_to_tem_body
--
Check template files...
OK: /Users/julien/code/spinalcordtoolbox/data/PAM50/template/PAM50_t2.nii.gz
OK: /Users/julien/code/spinalcordtoolbox/data/PAM50/template/PAM50_levels.nii.gz
OK: /Users/julien/code/spinalcordtoolbox/data/PAM50/template/PAM50_cord.nii.gz
Check parameters:
Data: megre3d_cr.nii.gz
Landmarks: megre3d_cr_label.nii.gz
Segmentation: megre3d_cr_msc.nii.gz
Path template: /Users/julien/code/spinalcordtoolbox/data/PAM50
Remove temp files: 1
Check input labels...
Creating temporary folder (/var/folders/5f/6n99hhyj5_g5cv_1bbx1q7cr0000gn/T/sct_2023-09-25_10-27-45_register-to-template_n_sjkkp7)
Copying input data to tmp folder and convert to nii...
Generate labels from template vertebral labeling
Check if provided labels are available in the template
ERROR: Wrong landmarks input. Labels must have correspondence in template space.
Label max provided: 99.0
Label max from template: 20
The program complains that the max value on the template is 20, even though I’ve edited the file to add labels 98 and 99. Proof:
**julien-macbook:**~/code/spinalcordtoolbox/data/PAM50/template $ sct_label_utils -i PAM50_label_body.nii.gz -display
--
Spinal Cord Toolbox (git-master-76c27e16f13974de3ff8f81e25c92abe8f6d80ec)
sct_label_utils -i PAM50_label_body.nii.gz -display
--
Position=(70,70,951) -- Value= 1
Position=(70,70,924) -- Value= 2
Position=(70,70,890) -- Value= 3
Position=(70,70,853) -- Value= 4
Position=(70,70,818) -- Value= 5
Position=(70,71,787) -- Value= 6
Position=(70,71,754) -- Value= 7
Position=(70,70,716) -- Value= 8
Position=(70,70,671) -- Value= 9
Position=(70,70,625) -- Value= 10
Position=(70,70,577) -- Value= 11
Position=(70,70,527) -- Value= 12
Position=(70,70,476) -- Value= 13
Position=(70,70,424) -- Value= 14
Position=(70,71,370) -- Value= 15
Position=(70,71,317) -- Value= 16
Position=(70,71,261) -- Value= 17
Position=(70,71,200) -- Value= 18
Position=(70,70,142) -- Value= 19
Position=(70,71,91) -- Value= 20
Position=(70,70,164) -- Value= 98
Position=(70,70,40) -- Value= 99
OK, I identified the cause of the problem. In fact, the file PAM50_label_body.nii.gz is not used by sct_register_to_template -l, but instead is generated on-the-fly from the file PAM50_levels.nii.gz. Issue opened: Confusing existence of `PAM50_label_body.nii.gz` · Issue #4229 · spinalcordtoolbox/spinalcordtoolbox · GitHub
Until this issue is resolved, I suggest to only use -ldisc with custom labels.
-ref subject
sct_register_to_template -i $anat -s $mask -ldisc $label -c t2 -ref subject -ofolder reg_to_sub
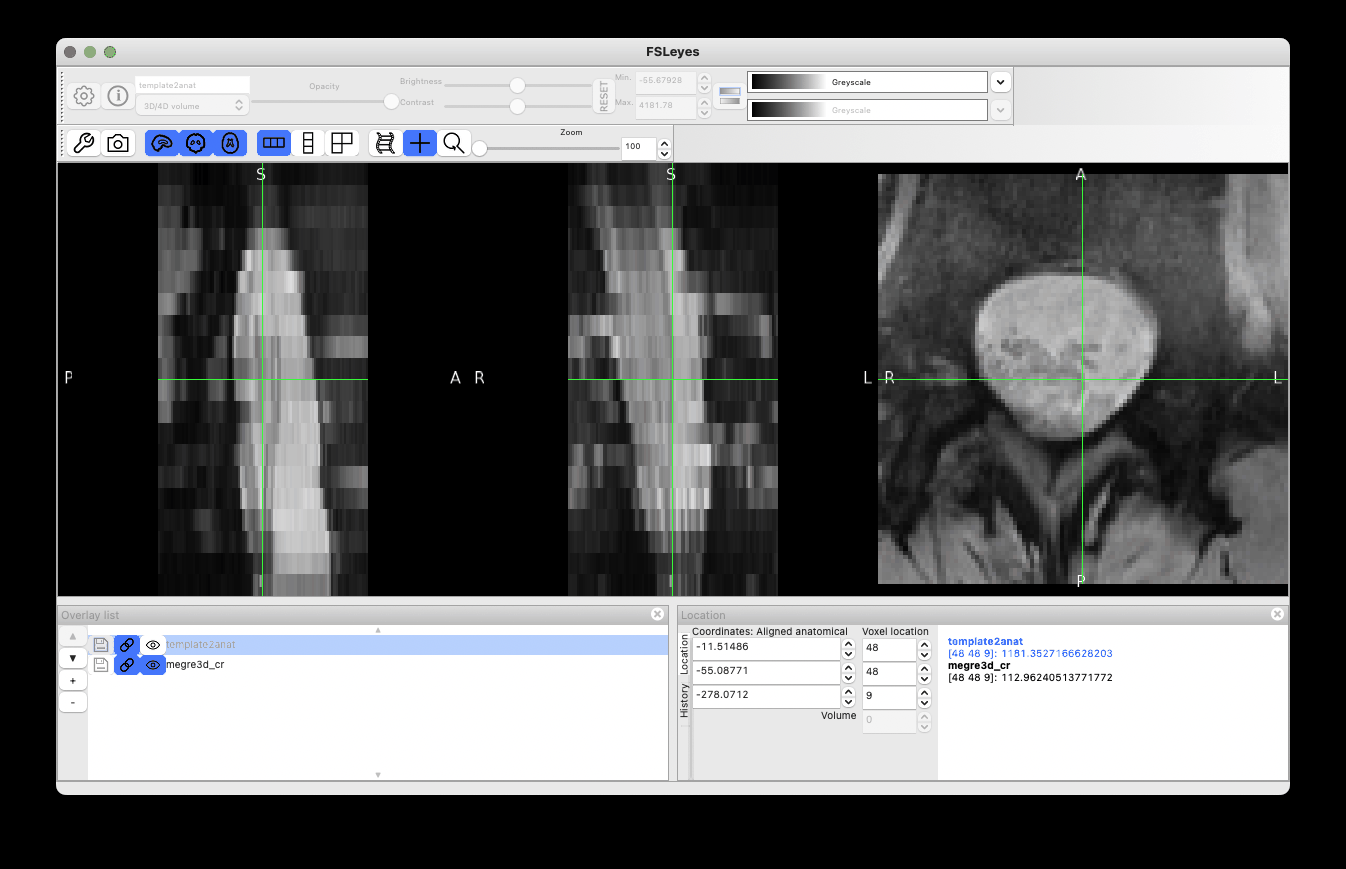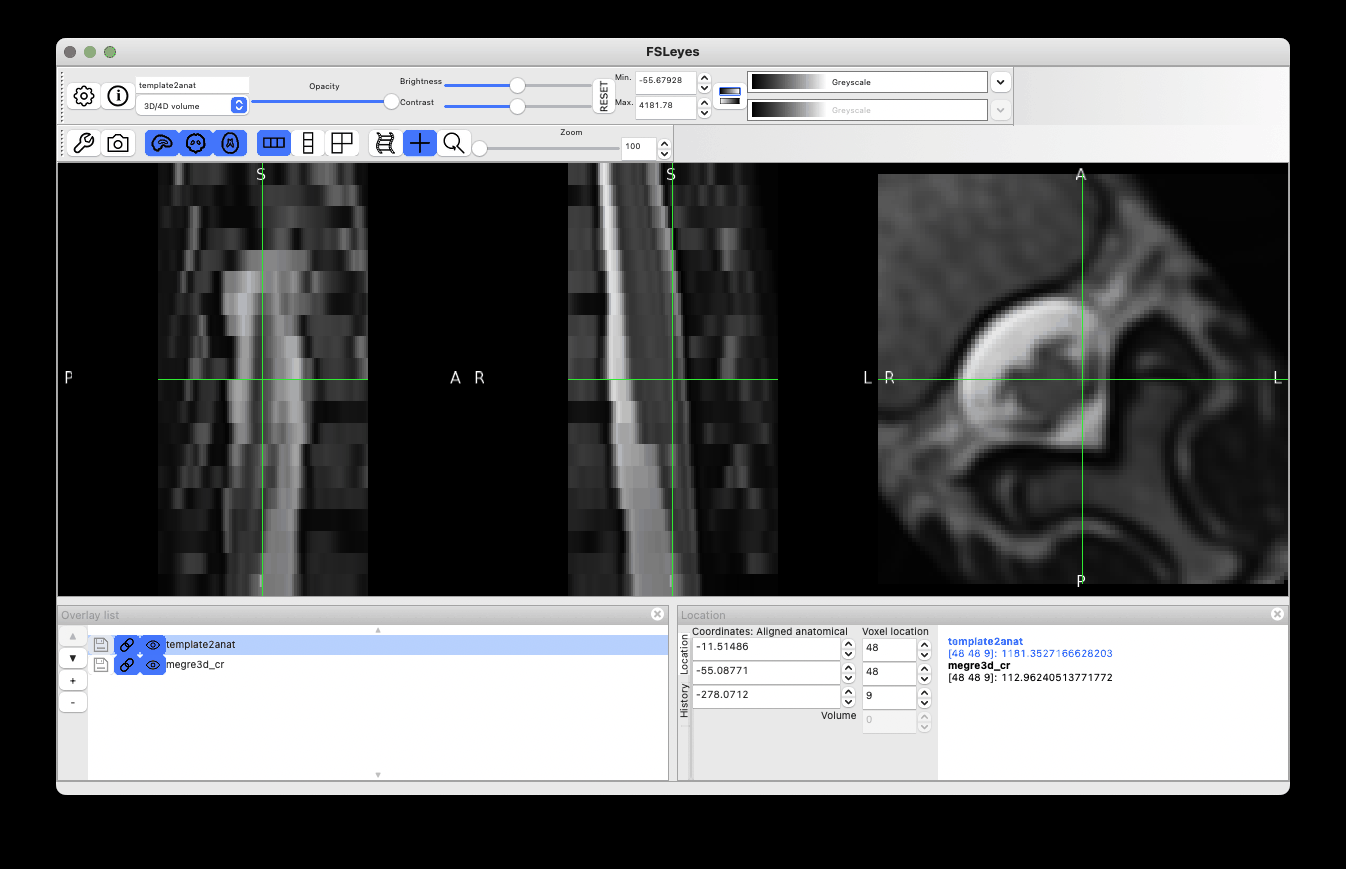
As you noted, there is definitely an issue with excessive rotation, which might be caused by a wrong estimation of spinal cord orientation at step=1. This is not uncommon in the lumbar area, where the spinal cord is rounder (vs. in the cervical cord, where the cord is more ellipsoid). To overcome this issue, I tried ignoring the rotation by using the centermass algorithm instead:
sct_register_to_template -i $anat -s $mask -ldisc $label -c t2 -ref subject -ofolder reg_to_sub -param step=1,type=seg,algo=centermass:step=2,type=seg,algo=bsplinesyn,metric=MeanSquares,iter=3,smooth=1 -qc qc -v 2 -r 0
However it did not solve the rotation problem. So I had to dig further, investigating the intermediate files and labels for registering data at step=0. Indeed, step=0 seems to be the cause of the issue:
The label registration at step=0 seems to be working fine:
So maybe the issue is caused by how labels are defined? Normally, one label orthogonal to the two provided labels is created, in order to properly estimate an affine transform. However, from the screenshot above, only two labels (not three) are used.
Digging further, I noticed that the third label uses a dummy value 99:
Which is problematic, because 99 is also the value chosen by the user (!). This is quite unfortunate 
So what we need to do:
- SCT dev team: add a check to make sure that the input image label does not have the value 99.
- For this particular case to work: modify the label value from 99–>97.
I replaced 99 by 97 in the following files:
- megre3d_cr_label.nii.gz
- PAM50_label_disc.nii.gz (which was previously modified by @kuech)
And then re-launched the registration:
sct_register_to_template -i $anat -s $mask -ldisc megre3d_cr_label_97-98.nii.gz -c t2 -ref subject -ofolder reg_to_sub -param step=1,type=seg,algo=centermass:step=2,type=seg,algo=bsplinesyn,metric=MeanSquares,iter=3,smooth=1 -qc qc -v 2 -r 0
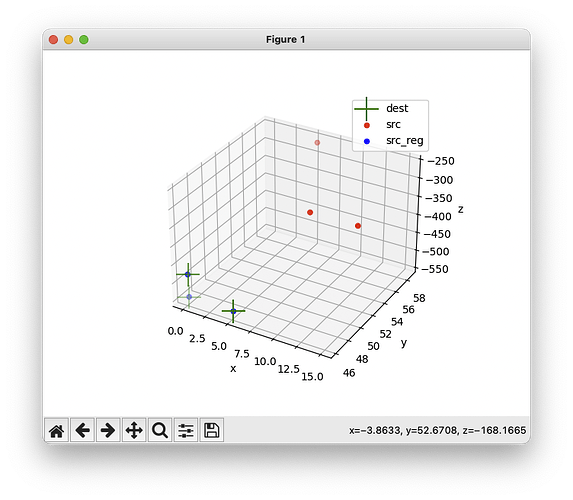
Bingo!  The rotation issue is now solved:
The rotation issue is now solved:
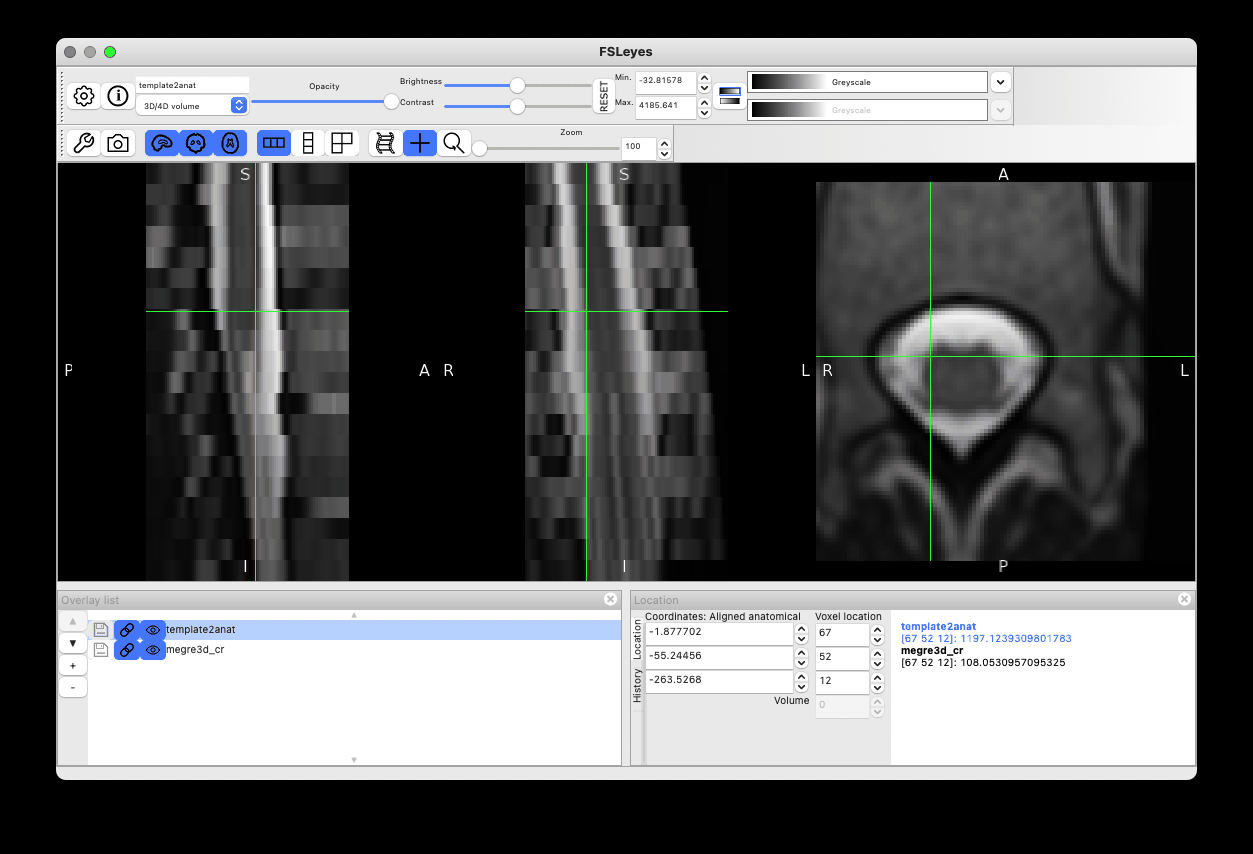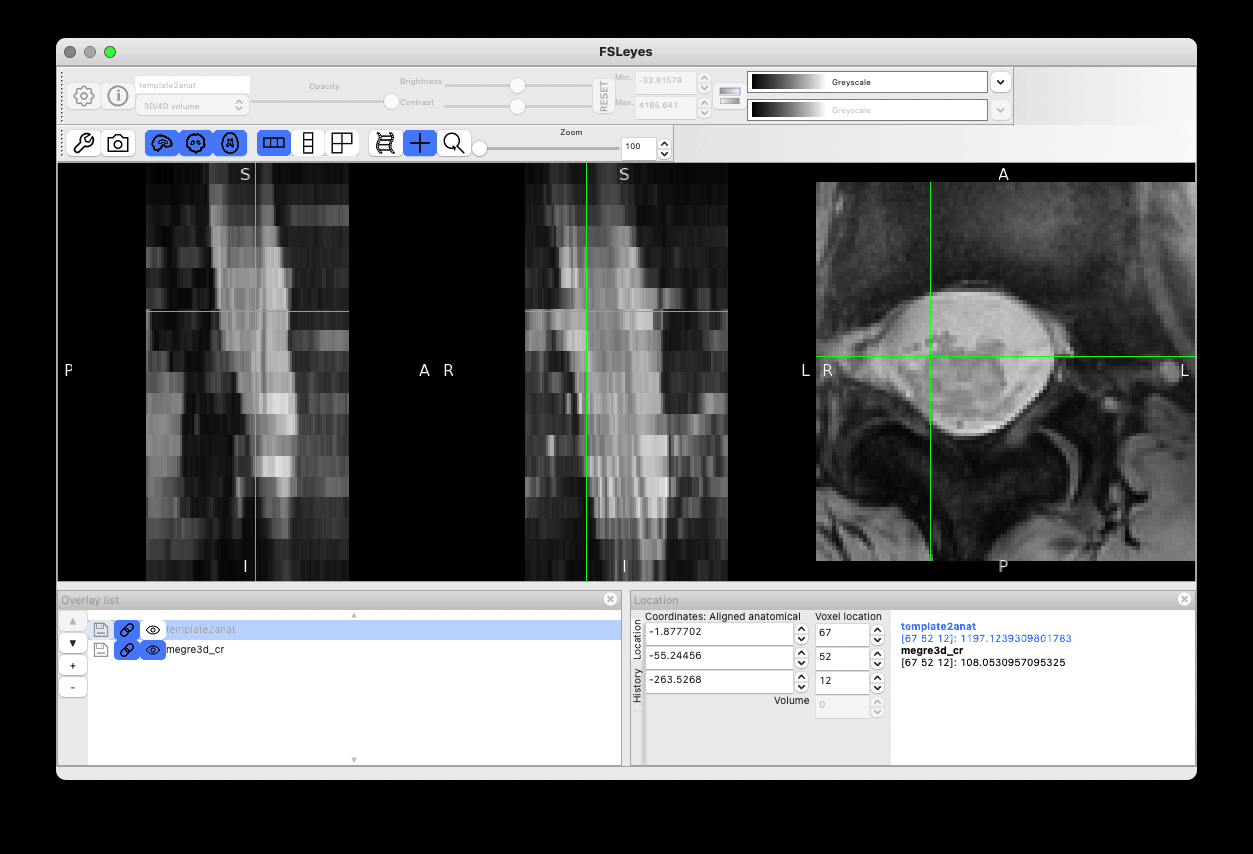
And the label-based registration at step=0 (output with -v 2) confirms there are now three labels:
Next step:
Things I’ve tried to improve registration:
| Parameters |
Results |
centermassrot vs. centermass |
 |
type=imseg,algo=centermassrot,rot_method=pcahog |
 |
-c t2s |
 (some slices from the T2s template are missing caudally) (some slices from the T2s template are missing caudally) |
step=3,type=im,algo=syn,metric=CC,iter=5,slicewise=1 |
 |
Suggested syntax:
sct_register_to_template -i $anat -s $mask -ldisc megre3d_cr_label_97-98.nii.gz -c t2 -ref subject -ofolder reg_to_sub -param step=1,type=imseg,algo=centermassrot,rot_method=pcahog:step=2,type=seg,algo=bsplinesyn,metric=MeanSquares,iter=3,smooth=1,slicewise=1:step=3,type=im,algo=syn,metric=CC,iter=5,slicewise=1 -qc qc
Results:

QC reports of all my investigations: qc.zip (877.1 KB)