Hi everyone,
I’m trying to apply segmentation algorithms to MRI lumbar images.
I’ve tried different combinations of commands and parameters, and the combination that seems to work best is the following:
sct_image -i filename -setorient LPI -o t2LPI.nii.gz
sct_deepseg_sc -i t2LPI.nii.gz -c t2 -kernel 3d -centerline cnn -brain 0 -o outputFilename.nii
sct_deepseg_sc -i t2LPI.nii.gz -c t2 -centerline file -file_centerline outputFilename.nii -brain 0 -o outputFilename2.nii
sct_create_mask -i t2LPI.nii.gz -p outputFilename2.nii -o mask.nii
sct_deepseg_sc -i t2LPI.nii.gz -c t2 -centerline file -file_centerline mask.nii -brain 0 -o outputFilename3.nii
sct_deepseg_sc -i t2LPI.nii.gz -c t2 -centerline file -file_centerline outputFilename3.nii -brain 0 -o outputFilename4.nii
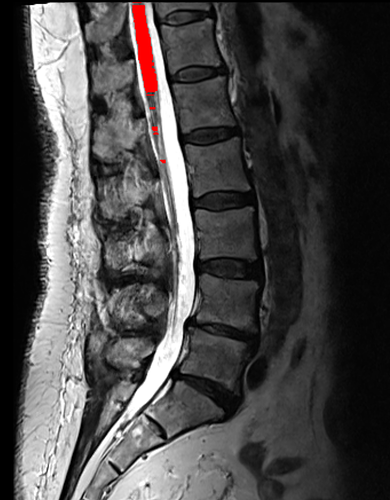
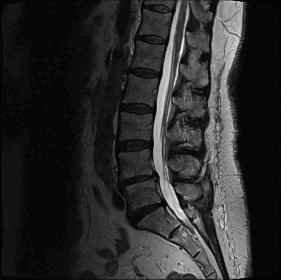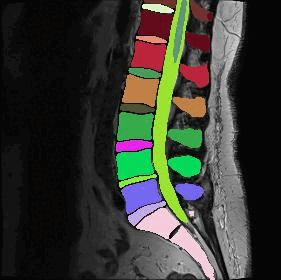
However, I am still getting poor segmentation results. Some non-spinal cord areas are being segmented, while the segmentation of the spinal cord itself is fragmented.
Indeed, I noticed that this approach only works on some images, while errors are raised for others (different errors depending on the image, such as the centerline not being detected using the CNN method). These images are NIfTI images that have been augmented on the z-axis using a Soup-GAN.
thank you for your help!