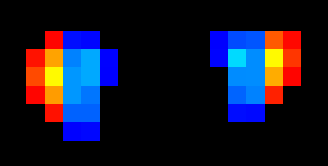Hi Julien,
I calculated WM-GM ratios for corticospinal tract for 10 subjects. I want to show that the medial half of the tract bundle is more affected than the lateral half of the lateral corticospinal tract bundle?
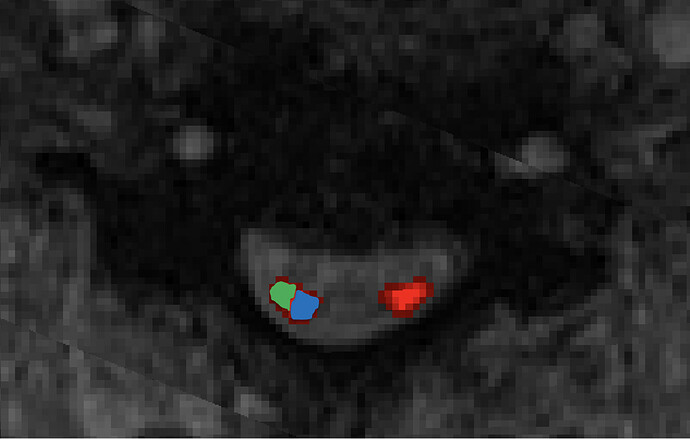
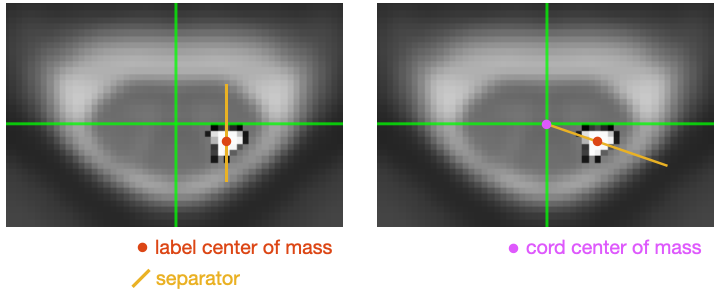
Is there a way to calculate metrics for medial 1/2 (blue) and lateral 1/2 (green) of the lateral corticospinal tracts separately -see figure for example?
Thanks
Hi @AdiVeeMiami
The key here is to split the right and left lateral CST label into two separate labels. Once the labels are split, they could be used by adding two entries into the atlas/info_label.txt file and call these entries with sct_extract_metric.
The splitting could easily be done (~1h of work) by designing a script that would split each label according to some rules. A question for you is: what rule would that be? Below are two suggestions. Let me know if either is acceptable:
Also, my time is quite limited at the moment, so if you are a bit familiar with Python and image processing, is something you would like to implement?
Cheers,
Julien
Hi Julien,
Thanks for quick response. The one on the left is what we need = ‘label center of mass’. This will divide the corticospinal tract into medial and lateral components.
I am not familiar with Python and image processing. I have started working with SCT recently. I would greatly appreciate your help with this script.
I am happy to email you directly regarding potential collaboration for your help with designing this script.
Thanks,
Aditya
OK no worries, we will create a script for you. I cannot guaranty when though. Do you have a specific timeline?
Thank you so much. Is April 15th a workable deadline? Thanks
Hi @AdiVeeMiami. I work with SCT, and have created a script for you to perform this task: split_labels.py (3.5 KB)
The usage for this script is as follows:
# Call the script using the python in SCT's virtual environment
$SCT_DIR/python/envs/venv_sct/bin/python split_labels.py -i label/atlas/PAM50_atlas_04.nii.gz label/atlas/PAM50_atlas_05.nii.gz
The values of the lateral/medial labels at the center voxels were weighted based on the position of the center of mass. The overlapping parts add to match the original CST label values:
To use the output files with ‘sct_extract_metric’, specify '-f' in the following way:
# Sample usage of the output files with 'sct_extract_metric'
sct_extract_metric -method map -i mtr.nii.gz -f PAM50_atlas_04_medial.nii.gz -o left-lcst_medial.csv
sct_extract_metric -method map -i mtr.nii.gz -f PAM50_atlas_04_lateral.nii.gz -o left-lcst_lateral.csv
sct_extract_metric -method map -i mtr.nii.gz -f PAM50_atlas_05_medial.nii.gz -o right-lcst_medial.csv
sct_extract_metric -method map -i mtr.nii.gz -f PAM50_atlas_05_lateral.nii.gz -o right-lcst_lateral.csv
Note: For ‘-f’ in ‘sct_extract_metric’, we specify the file name of the label (and not the “data/atlas” directory). This is slightly different than what was originally suggested by @jcohenadad in an earlier comment: Measure data for corticospinal tract - #2 by jcohenadad)
Please let me know if this works for your needs. 
1 Like
Hi Joshua,
Thanks for the script. I am new to SCT and python.
I downloaded the script- should I move the script to the SCT folder: $SCT_DIR/python/envs/venv_sct/bin/python
I actually don’t have a python folder in the bin folder.
Not sure how to proceed.
Thanks
I actually don’t have a python folder in the bin folder.
Hi @AdiVeeMiami. My apologies for the confusion – in this case, “python” is not a folder. “python” is a command.
So, when you see this line:
$SCT_DIR/python/envs/venv_sct/bin/python split_labels.py -i label/atlas/PAM50_atlas_04.nii.gz label/atlas/PAM50_atlas_05.nii.gz
You can understand the meaning like this:
python split_labels.py -i label/atlas/PAM50_atlas_04.nii.gz label/atlas/PAM50_atlas_05.nii.gz
In other words: We want to use ‘python’ from the directory ‘$SCT_DIR/python/envs/venv_sct/bin/’ to execute the script ‘split_labels.py’.
I downloaded the script- should I move the script
You should be able to run the script from any location. The only thing to keep in mind is that when you specify the path to your data files using ‘-i’, the paths will need to be relative to whichever location the script is run from.
1 Like
Thanks for the response.
Am not sure what I’m doing wrong but I am getting this response:
/python/envs/venv_sct/bin/python: can’t open file ‘split_labels.py’: [Errno 2] No such file or directory
Ah, my apologies. I may have said something misleading. You just need to make sure that your current working directory contains the ‘split_labels.py’ script.
Here is some example usage:
# Check to see that current working directory has 'split_labels.py'
❯ ls
label split_labels.py
# Check to see that the required files are present. I have them stored in the
# "label/atlas" folder in my working directory, but it may be different for you.
❯ ls label
atlas
❯ ls label/atlas
PAM50_atlas_04.nii.gz PAM50_atlas_05.nii.gz
# Run the command
❯ $SCT_DIR/python/envs/venv_sct/bin/python split_labels.py -i label/atlas/PAM50_atlas_04.nii.gz label/atlas/PAM50_atlas_05.nii.gz
Splitting label/atlas/PAM50_atlas_04.nii.gz into lateral and medial halves.
Saving generated files...
Splitting label/atlas/PAM50_atlas_05.nii.gz into lateral and medial halves.
Saving generated files...
1 Like
I put the file in the working directory. This is the error message I am getting:
File “split_labels.py”, line 13, in
from spinalcordtoolbox.utils import extract_fname
ImportError: cannot import name ‘extract_fname’
My apologies for the trouble. I wrote the script using the most recent version of SCT (v5.2.1) but I may need to make a small modification to the script if you are using an older version of SCT.
I have made an adjustment to the script so that it should work regardless of which SCT version is used.
Could you please try again with this new script? split_labels.py (3.6 KB)
Thanks for this. It works! Was able to process my data.
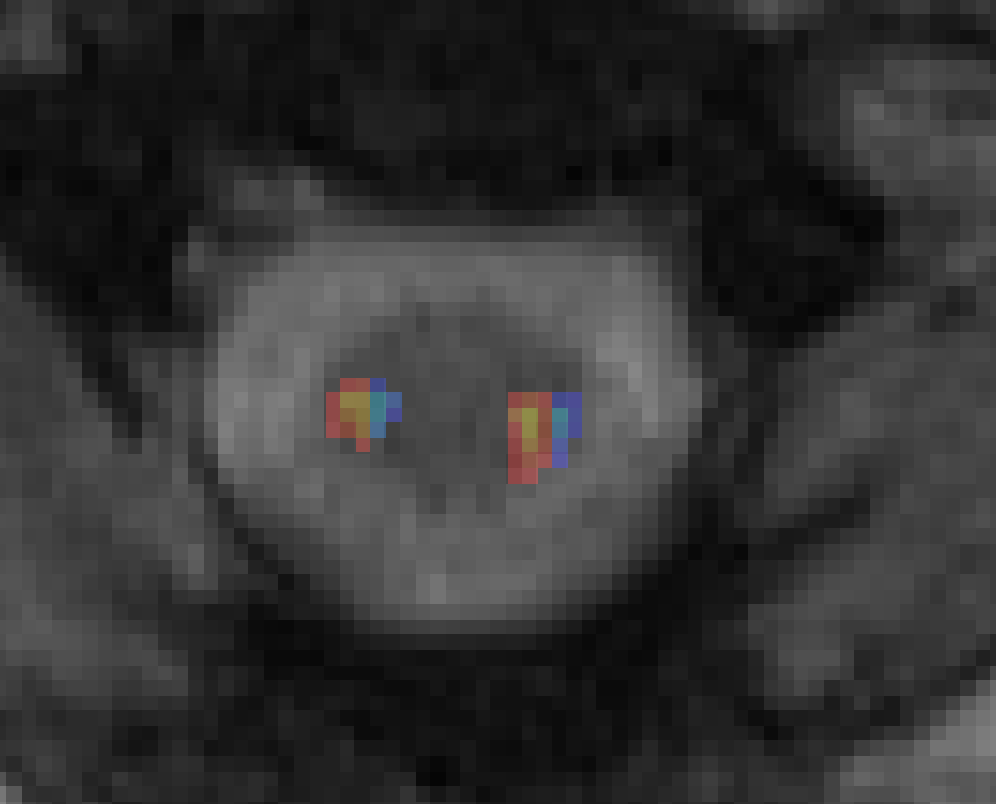
I did find that the medial and lateral switched for the left Lateral CST. Here are two screenshots. I assigned the lateral half to yellow and the medial half to blue. On the right side- it is correct. But on the left side- it is switched. I made the adjustment to my data.
Ah! You are absolutely correct. This was a mistake on my part: I mistook the definitions of “medial” and “lateral”, and instead both images are split into “left” and “right” halves (as you have pointed out).
My sincere apologies for my misunderstanding. I have updated the script once more: split_labels.py (4.1 KB)
The usage is now as follows:
❯ $SCT_DIR/python/envs/venv_sct/bin/python split_labels.py -i-left label/atlas/PAM50_atlas_04.nii.gz -i-right label/atlas/PAM50_atlas_05.nii.gz
(This way, the left/right tracts are explicitly specified using ‘-i-left’ and ‘-i-right’)
Now it should correctly split the left-side image.
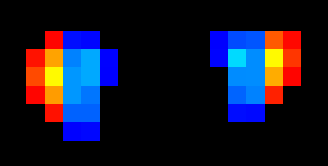
Thanks for this! Once again, grateful for your help!
1 Like