Hello SC toolbox team:
I’m trying to register two spinal cord MRI images of the same person in different directions to the PAM50 template.
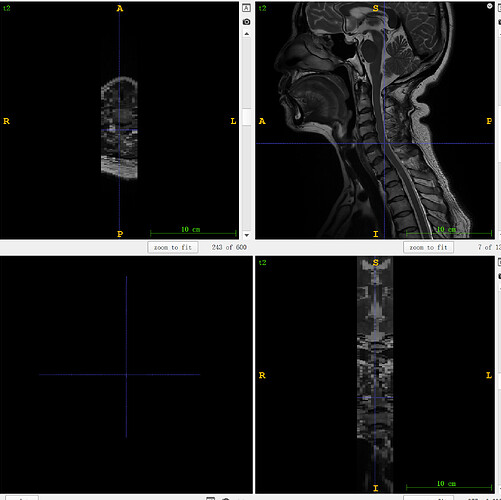
t2:
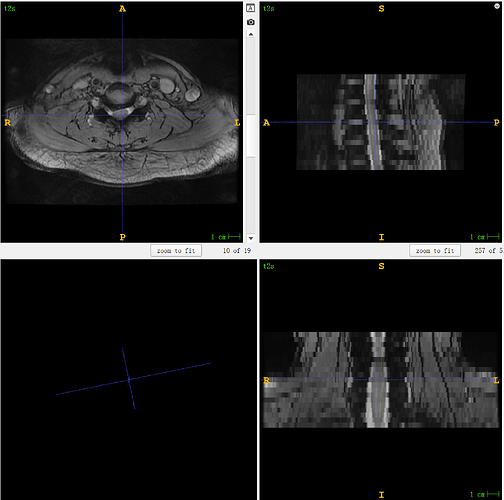
t2s:
I use these command
sct_propseg -i t2.nii.gz -c t2 -qc ~/qc_singleSubj
sct_label_vertebrae -i t2.nii.gz -s t2_seg.nii.gz -c t2 -qc ~/qc_singleSubj
sct_label_utils -i t2_seg_labeled.nii.gz -vert-body 3,6 -o t2_labels_vert.nii.gz
sct_register_multimodal -i t2s.nii.gz -d t2.nii.gz -identity 1 -x nn
sct_apply_transfo -i t2s.nii.gz -d t2.nii.gz -w warp_t2s2t2.nii.gz -o t2s_t2.nii.gz
sct_register_to_template -i t2.nii.gz -s t2_seg.nii.gz -l t2_labels_vert.nii.gz -c t2 -qc ~/qc_singleSubj
sct_apply_transfo -i t2s_t2.nii.gz -d anat2template.nii.gz -w warp_anat2template.nii.gz -o t2s_template.nii.gz
Is this progress right?
Thank you for your time!
Hi @janessa
Almost. The only differences are that
- I suggest to register the T2s to the PAM50 using the intermediate T2->PAM50 transfo, instead of registering
- I recommend to also segment the SC on the T2s scan, to ensure better registration
- I recommend switching from propseg to deepseg_sc
This is similar to what is done in the batch_processing.sh example script.
So it would look like this:
# Segment T2
sct_deepseg_sc -i t2.nii.gz -c t2 -qc ~/qc_singleSubj
# Label T2 vertebrae
sct_label_vertebrae -i t2.nii.gz -s t2_seg.nii.gz -c t2 -qc ~/qc_singleSubj
sct_label_utils -i t2_seg_labeled.nii.gz -vert-body 3,6 -o t2_labels_vert.nii.gz
# Register T2 to PAM50
sct_register_to_template -i t2.nii.gz -s t2_seg.nii.gz -l t2_labels_vert.nii.gz -c t2 -qc ~/qc_singleSubj
# Segment T2s
sct_deepseg_sc -i t2s.nii.gz -c t2s -qc ~/qc_singleSubj
# Register PAM50 to T2s
sct_register_multimodal -i ${SCT_DIR}/data/PAM50/template/PAM50_t2s.nii.gz -iseg ${SCT_DIR}/data/PAM50/template/PAM50_cord.nii.gz -d t2s.nii.gz -dseg t2s_seg.nii.gz -param step=1,type=seg,algo=centermass:step=2,type=seg,algo=bsplinesyn,slicewise=1,iter=3:step=3,type=im,algo=syn,slicewise=1,iter=1,metric=CC -initwarp warp_template2anat.nii.gz -initwarpinv warp_anat2template.nii.gz -owarp warp_template2t2s.nii.gz -owarpinv warp_t2s2template.nii.gz
Hope that helps!
Thank you for the quick response! This is very helpful. But I have another question. I wonder if i can use sct to get the image just include the partial spine, like C3-C6, from t2 and t2s.
Yes, you can crop the image to only keep specific levels, or you can compute cord morphometrics or extract qMRI metric (DTI, MT etc.) across specific levels, etc. These scenarios are well described in the SCT tutorials. I suggest you do these
tutorials, and if you have specific questions please feel free to ask.
Hi,
Thanks for the pointers.This process work well for many images,but some MRI images show artifacts after being registered to the PAM50 template. What could be the reason for this? Do you have any suggestions to solve this problem?
Thank you for your time!
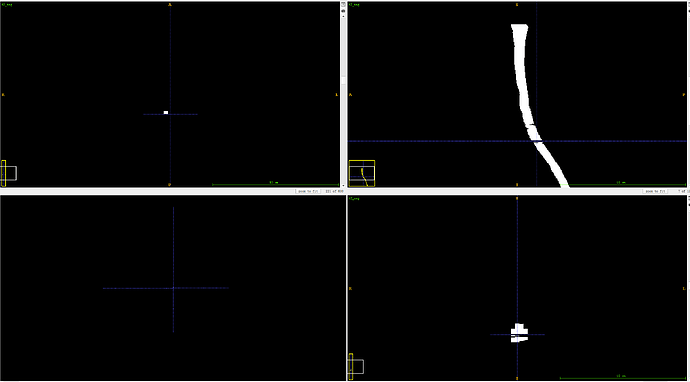
Issue is likely related to an inaccurate spinal cord segmentation. Can you upload your segmentation (in nii.gz format)?
t2_seg.nii.gz (26.5 KB)
You are right.There are some slice missing.Do you have any suggestions to solve this problem?