Hello everyone,
I have been working with the sct_register_multimodal tool and experimenting with different parameter settings for image registration between T1-weighted and fMRI data. Specifically, I tried the following configurations:
-
-param step=1, type=im, algo=dl
-
-param step=1, type=seg, algo=slicereg: step=2, type=seg, algo=centermass, slicewise=1, iter=3
I observed that the second configuration seems to yield better results. However, I wanted to ask the community: Are there any other recommended or optimal parameter settings that might produce even better results than what I’ve tried so far?
Here is my data
fmri_spinal_moco_mean_seg.nii.gz (2.9 KB)
fmri_spinal_moco_mean.nii.gz (3.7 MB)
t1_spinal.nii.gz (6.4 MB)
Thanks!!!
Hi,
Here is my QC report: qc.zip (941.3 KB)
Hi Julien,
Thanks for your detailed explanation and support. Under your guidance, most of the data achieved very good registration results, which has been extremely helpful.
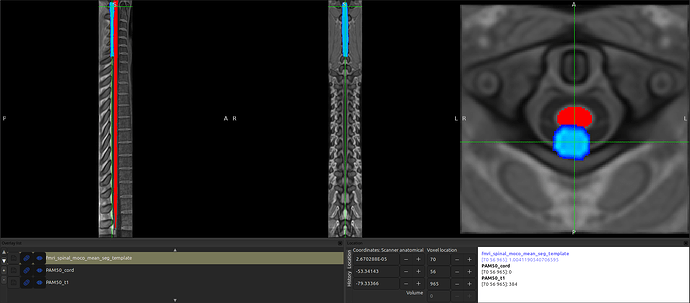
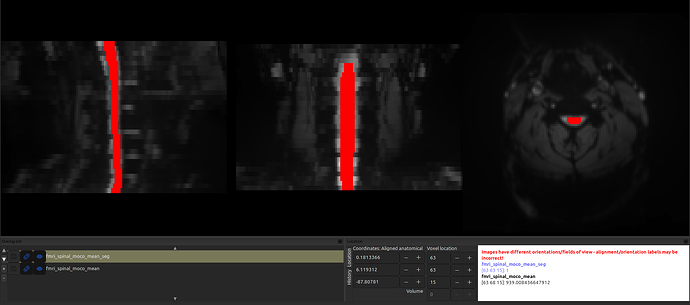
However, I have encountered an issue with a few data. After registering the fMRI data to the template, I noticed a shift, as shown in the image (and in the data).
Upon further inspection, I found that there is no issue when registering T1 to fMRI, nor is there any problem when registering T1 to the template. The problem arises when applying the transformation from fMRI to the template, which leads to the observed shift.
sct_apply_transfo -i ${datadir}/${sub}/spinal/fmri_spinal_moco_mean_seg.nii.gz \
-d ${template} \
-w ${datadir}/${sub}/spinal/${base_out}/warp_fmri_spinal_moco_mean2t1_spinal.nii.gz ${datadir}/${sub}/t1/spinal/reg/warp_anat2template.nii.gz\
-o ${datadir}/${sub}/spinal/${base_out}/fmri_spinal_moco_mean_seg_template.nii.gz
I have tried different interpolation methods, but it seems that none of them resolve the issue. I would greatly appreciate your suggestions on how to address this problem. Thank you again for your help.
Best,
Xiaomin
I was able to reproduce the issue you describe.
For posterity, I have updated the commands to use the exact paths found in the sub.zip “data” link that you shared (to help @jcohenadad debug);
frmi → t1 → template
cd sub/sub # extracted folder
sct_apply_transfo -i fmri_spinal_moco_mean_seg.nii.gz \
-d ${SCT_DIR}/data/PAM50/template/PAM50_cord.nii.gz \
-w reg_test/warp_fmri_spinal_moco_mean2t1_spinal.nii.gz t1/reg/warp_anat2template.nii.gz\
-o fmri_spinal_moco_mean_seg_template.nii.gz
template → t1 → fmri
cd sub/sub # extracted folder
sct_apply_transfo -i ${SCT_DIR}/data/PAM50/template/PAM50_cord.nii.gz \
-d fmri_spinal_moco_mean_seg.nii.gz \
-w t1/reg/warp_template2anat.nii.gz reg_test/warp_t1_spinal2fmri_spinal_moco_mean.nii.gz \
-o PAM50_cord-to-fmri.nii.gz
I will continue to try to debug. I just wanted to quickly share this post to save time with reproducing the issue.
Kind regards,
Joshua
Thank you so much Joshua!
Apologies for the late reply! I had a little more time to look at this now. 
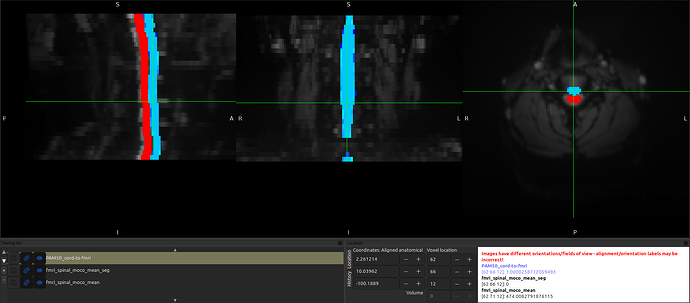
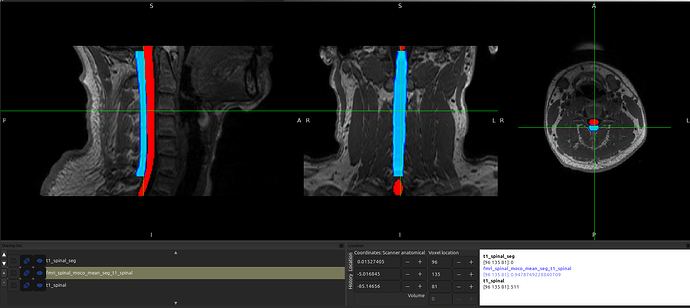
Focusing in on just the “fMRI to T1” registration, I see the same shift when warping the fMRI image to the T1 space by running the following command:
sct_apply_transfo -i fmri_spinal_moco_mean_seg.nii.gz \
-d t1/t1_spinal.nii.gz \
-w reg_test/warp_fmri_spinal_moco_mean2t1_spinal.nii.gz \
-o fmri_spinal_moco_mean_seg_t1_spinal.nii.gz
When I double-check the fMRI-T1 registration results by running the same command, just on the anat image instead of the segmentation, it’s fine?
sct_apply_transfo -i fmri_spinal_moco_mean.nii.gz \
-d t1/t1_spinal.nii.gz \
-w reg_test_2/warp_fmri_spinal_moco_mean2t1_spinal.nii.gz \
-o fmri_spinal_moco_mean_seg_t1_spinal.nii.gz
So, if the anat image can be warped OK, then something seems to be wrong with the _seg image specifically.
When I check the headers for both the original and the _seg image, I notice that the sform of the segmentation is shifted!
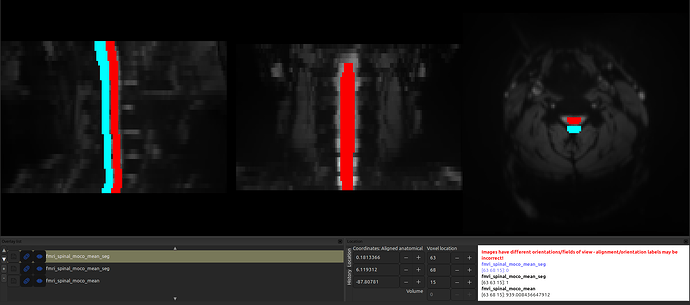
However, even though the sform is shifted, the segmentation is aligned with the anat image:
And, likewise, if I “fix” the sform of the segmentation (so that they are in the same space) by running the command sct_image -set-sform-to-qform -i fmri_spinal_moco_mean_seg.nii.gz, the shift appears!
So, tl;dr:
- The segmentation is not in the same space as the anat image (due to different sforms).
- Fixing the sform (but leaving the data array as-is) doesn’t fix the problem, since both the sform and the data array have this shift.
@Xiaomin_Lin, how was the segmentation generated?
Hi Joshua,
I just wanted to sincerely thank you for your help with the debugging! I took a look using Nibabel and, sure enough, I found that the data with the shift have discrepancies between their _seg sform and the original sform. I’m still not entirely sure how this happened, but I suspect it might be related to some weirdness that occurred when I manually modified the SEG file.
In any case, I really appreciate your assistance. Without your suggestion, I would never have thought to check this! Thanks again for pointing me in the right direction.
Best,
Xiaomin
1 Like
Thank you for such kind words! I’m very happy to help, and I appreciate your continued participation on the forum. 
I’m glad it helped! For future reference, you can use the command sct_image -header to check this information. (We created this flag as a mirror of the “fslhd” utility, so that users wouldn’t have to install the entirety of FSL just to check the header metadata. It’s a very useful tool!)
Anyway, feel free to continue to open new forum posts any time you have questions or concerns, and we’ll be happy to assist. 
Kind regards,
Joshua