Hi,
I’m analysing spinal MRI images for the first time and ran into something that might not be a problem at all but that I wanted to check to be sure.
SCT version: 7.0 OS: macOS
Pipeline:
- T1 processing:
sct_register_to_template→sct_warp_template - T2s processing:
sct_register_multimodal(PAM50_t2 → T2s, initialised with T1 warp via-initwarp) →sct_warp_template(T2s space) - fMRI processing:
sct_register_multimodalusing warpedPAM50_t2.nii.gzas source image →sct_warp_template(fMRI space)
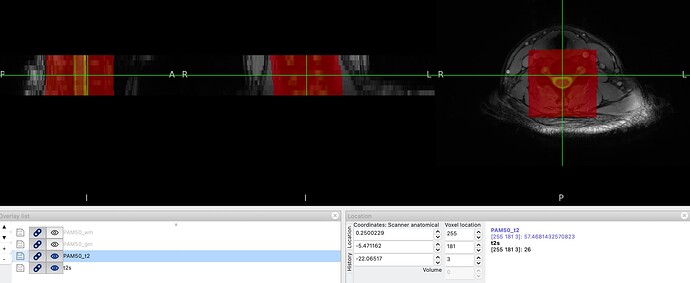
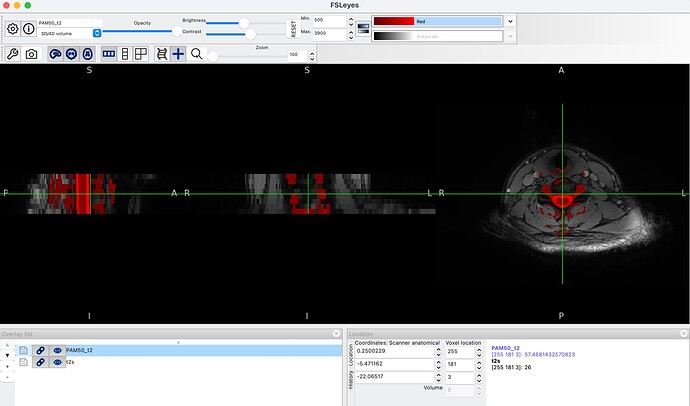
When viewing the original PAM50_t2 template, it contains substantial signal outside the spinal cord itself - vertebral bodies and surrounding tissue are clearly visible. After warping to subject T2s space, these surrounding structures are also present in the warped template.
In our fMRI pipeline, we use the warped PAM50_t2.nii.gz as the source image for registration to fMRI space:
bash:
sct_register_multimodal -i PAM50_t2.nii.gz \
-d fmri_mean.nii.gz \
-iseg PAM50_cord.nii.gz \
-dseg fmri_seg.nii.gz \
-param step=1,type=seg,algo=affine,slicewise=1,metric=MeanSquares,smooth=3:step=2,type=seg,algo=syn,metric=MeanSquares,smooth=1,iter=5
Questions
-
Does the presence of surrounding anatomy in
PAM50_t2.nii.gznegatively affect the quality of registration to fMRI space? -
Would it be better to mask
PAM50_t2.nii.gzwith the cord segmentation (PAM50_cord.nii.gz) before using it as a registration source, so that only cord signal is used for matching?