Hi Julian,
Sorry about the delay. Here are the results of my investigations.
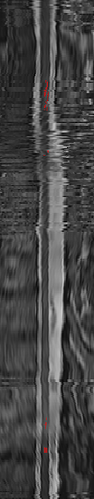
Firstly, it appears that the jittering is caused by the cord straightening procedure, which is part of sct_register_to_template. The straightened cord is output with the file name straight_ref.nii.gz, so that’s the one I compared across various experiments below.
Image resolution
I was wondering if highly anisotropic axial resolution (0.6x0.6x4.8mm) was causing that problem, so I resampled the image, segmentation and labels:
# Resample anat to 1mm iso
sct_resample -i sub-m023917_ses-20160302_acq-ax_T2w.nii.gz -mm 1 -x linear
# Change input segmentation type to FLOAT32 before resampling to linear for better precision
sct_image -i sub-m023917_ses-20160302_acq-ax_seg_T2w.nii.gz -type float32 -o sub-m023917_ses-20160302_acq-ax_seg_T2w_float32.nii.gz
# Resample seg to 1mm
sct_resample -i sub-m023917_ses-20160302_acq-ax_seg_T2w_float32.nii.gz -mm 1 -x linear
# Resample labels to 1mm
# Note: We cannot use simple nearest neighbour interpolation, otherwise labels will be duplicated with upsampling. Hence, we need to use `sct_apply_transfo`, which uses the center of mass of each label.
sct_register_multimodal -i sub-m023917_ses-20160302_acq-ax_disks_T2w.nii.gz -d sub-m023917_ses-20160302_acq-ax_T2w_r.nii.gz -identity 1
sct_apply_transfo -i sub-m023917_ses-20160302_acq-ax_disks_T2w.nii.gz -d sub-m023917_ses-20160302_acq-ax_T2w_r.nii.gz -w warp_sub-m023917_ses-20160302_acq-ax_disks_T2w2sub-m023917_ses-20160302_acq-ax_T2w_r.nii.gz -x label
However, when comparing straight_ref.nii.gz with/without resampling, the jittering is still there:

Quality of segmentation
Another concern is the quality of the segmentation. I noticed strange effects, for example the same segmentation across two adjacent slices, with sub-optimal matching between the cord and the mask:
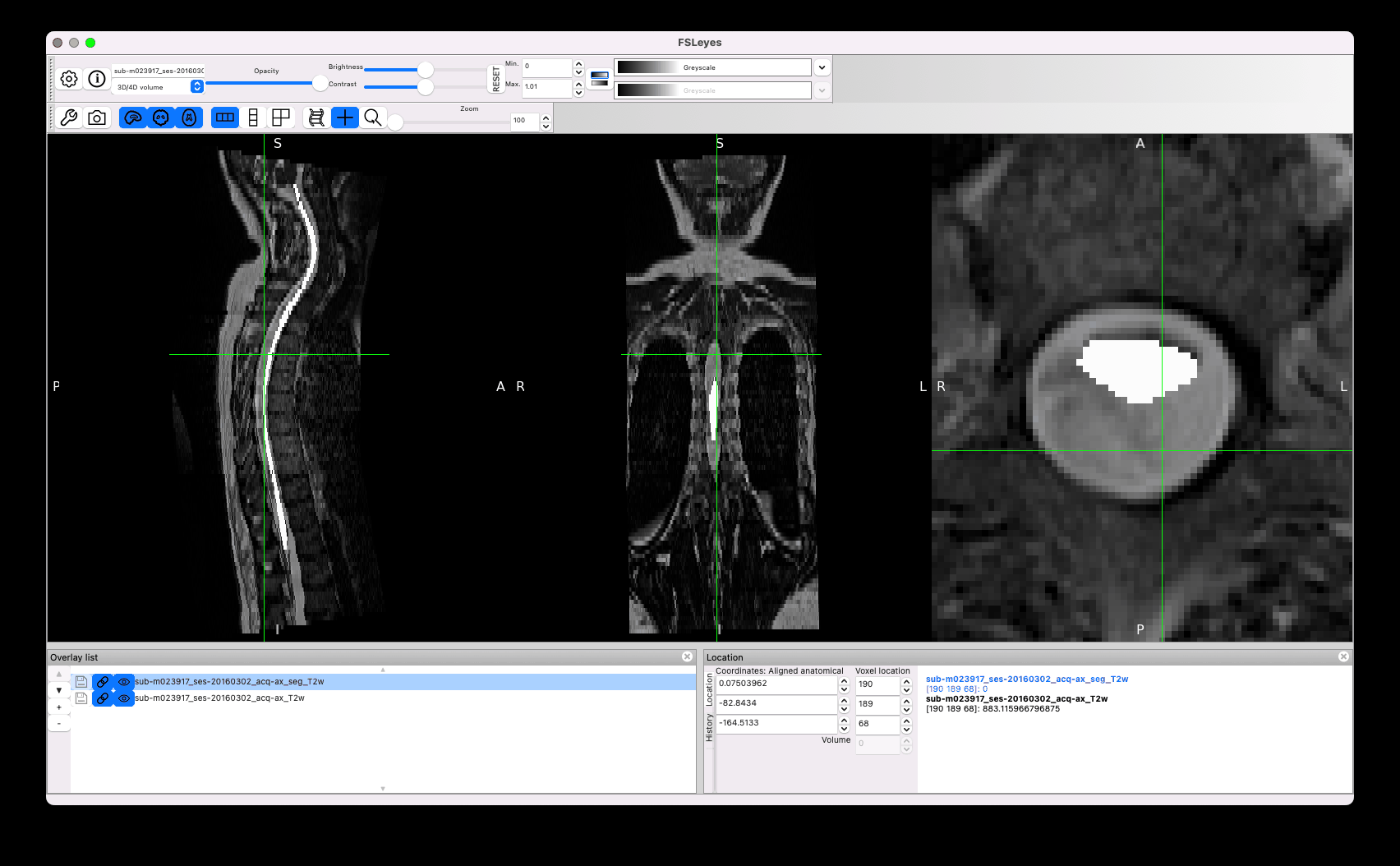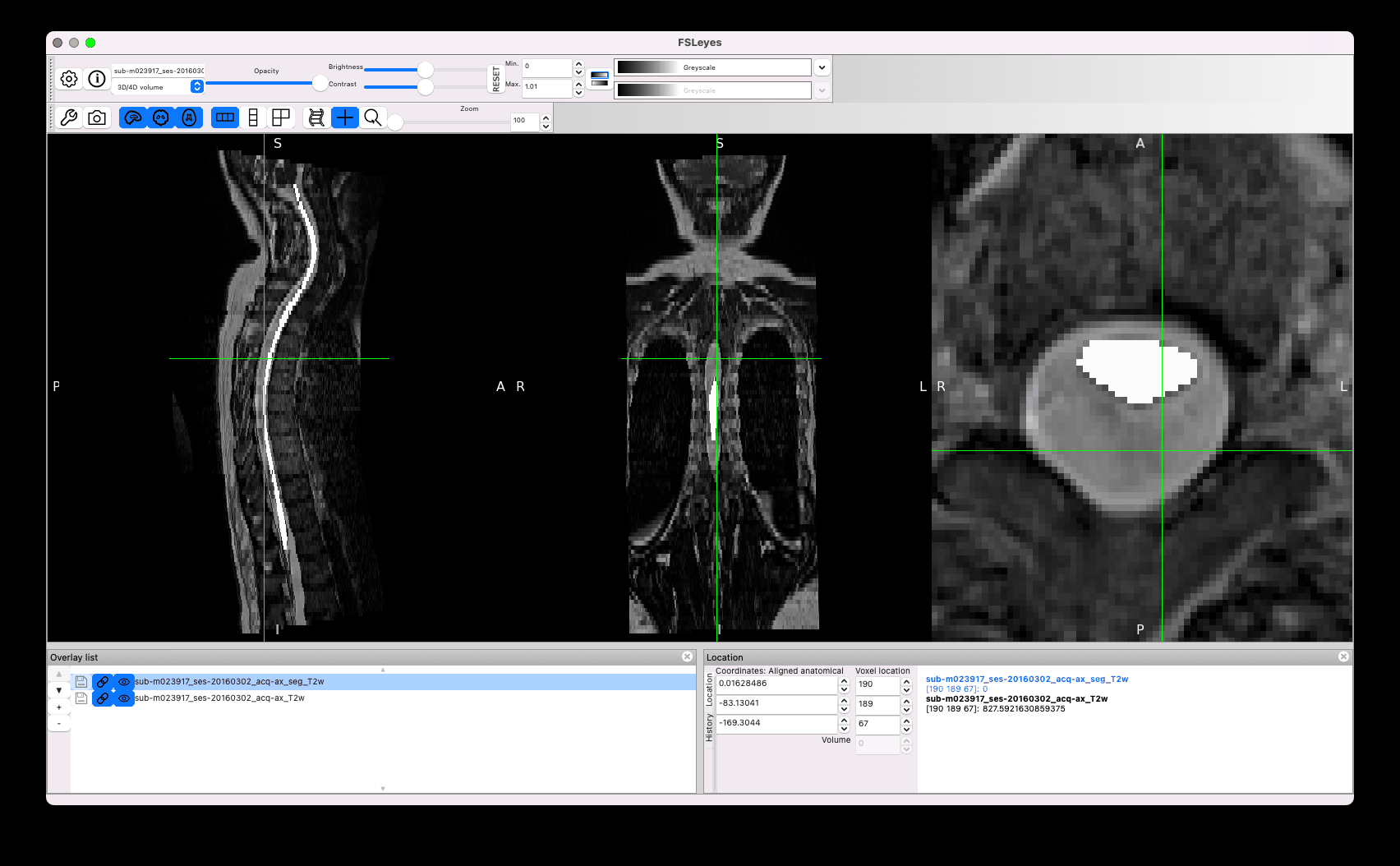
Other issues concern sub-optimal segmentation, such as the leaking below (the manual segmentation you provided is in white):
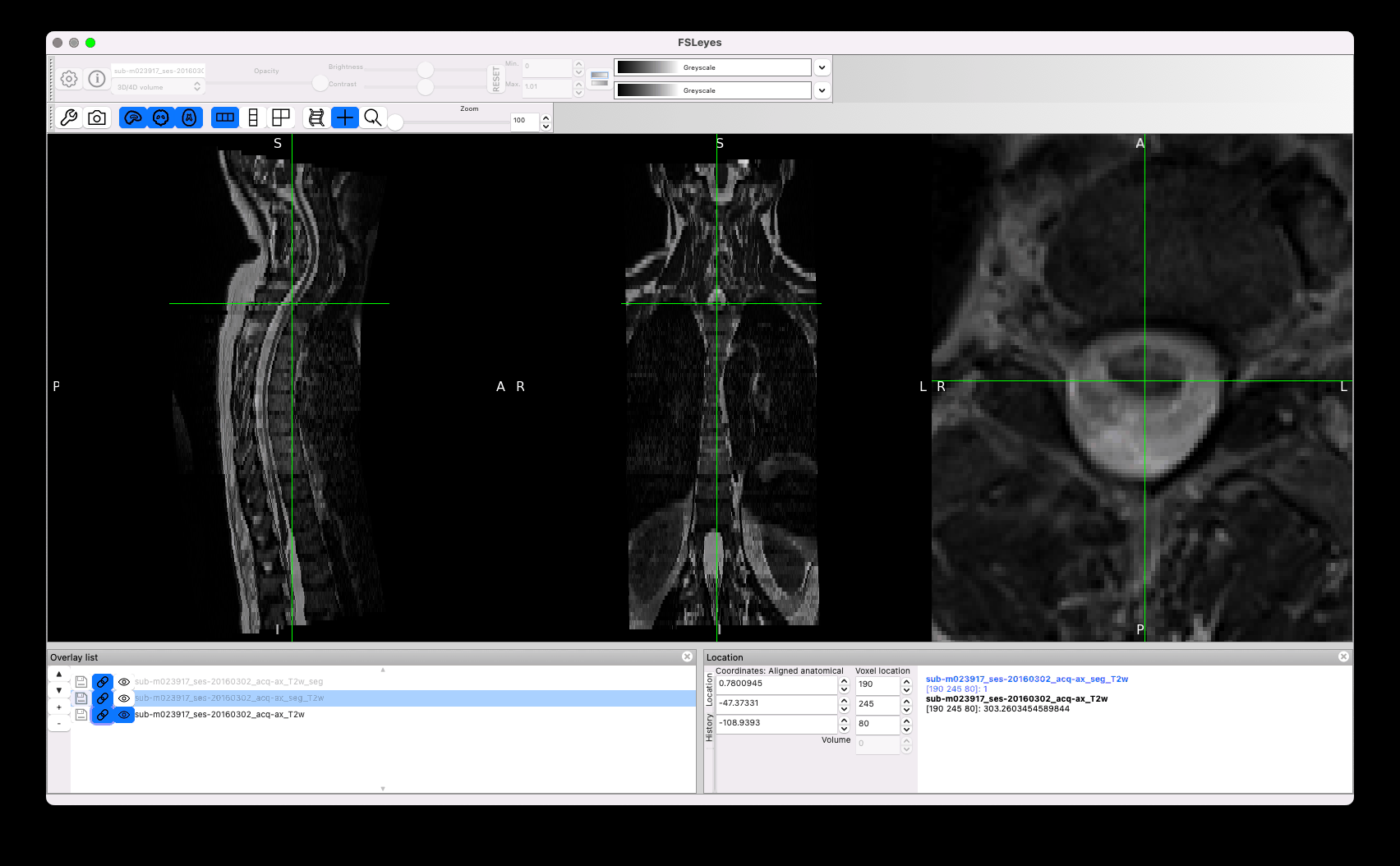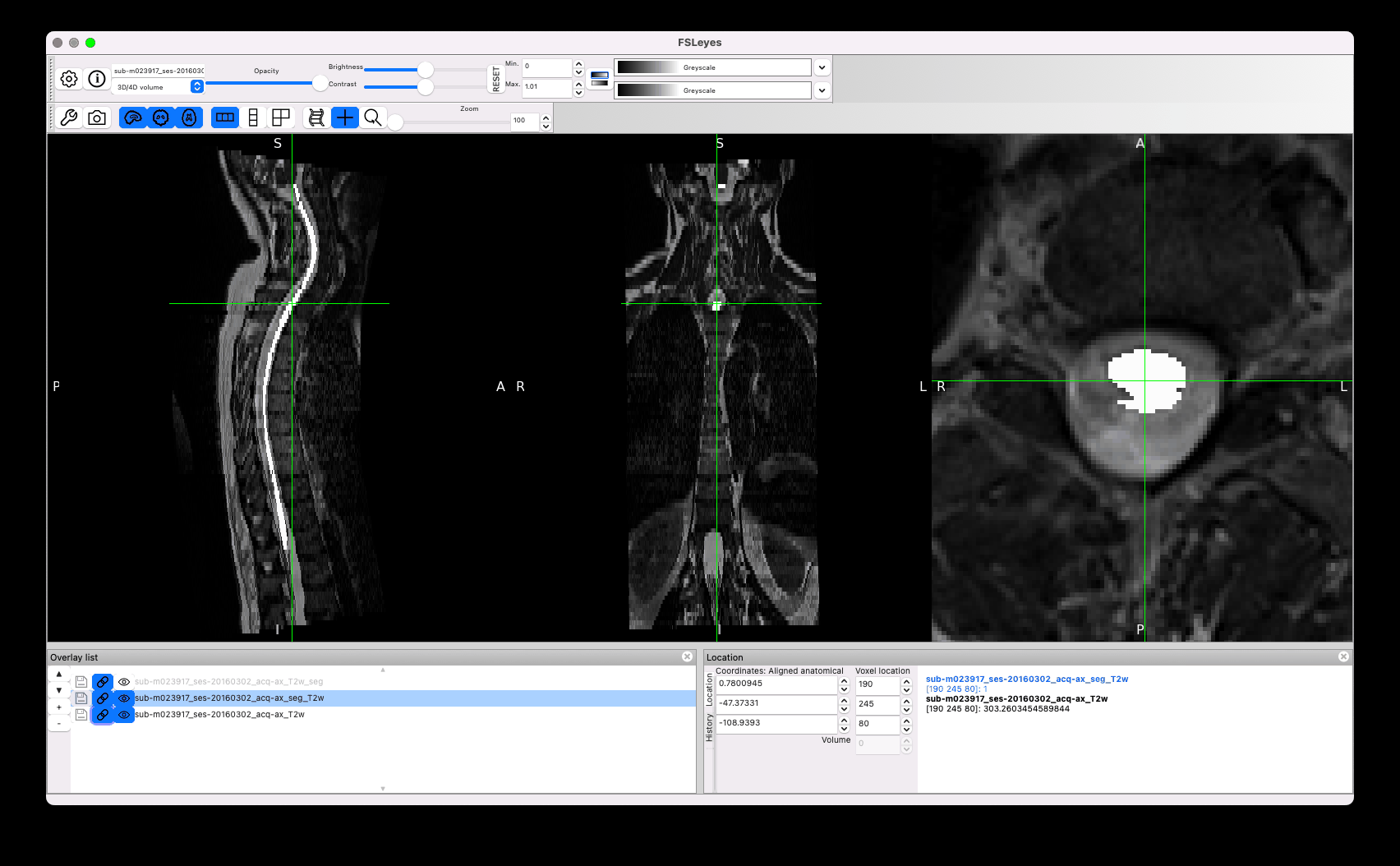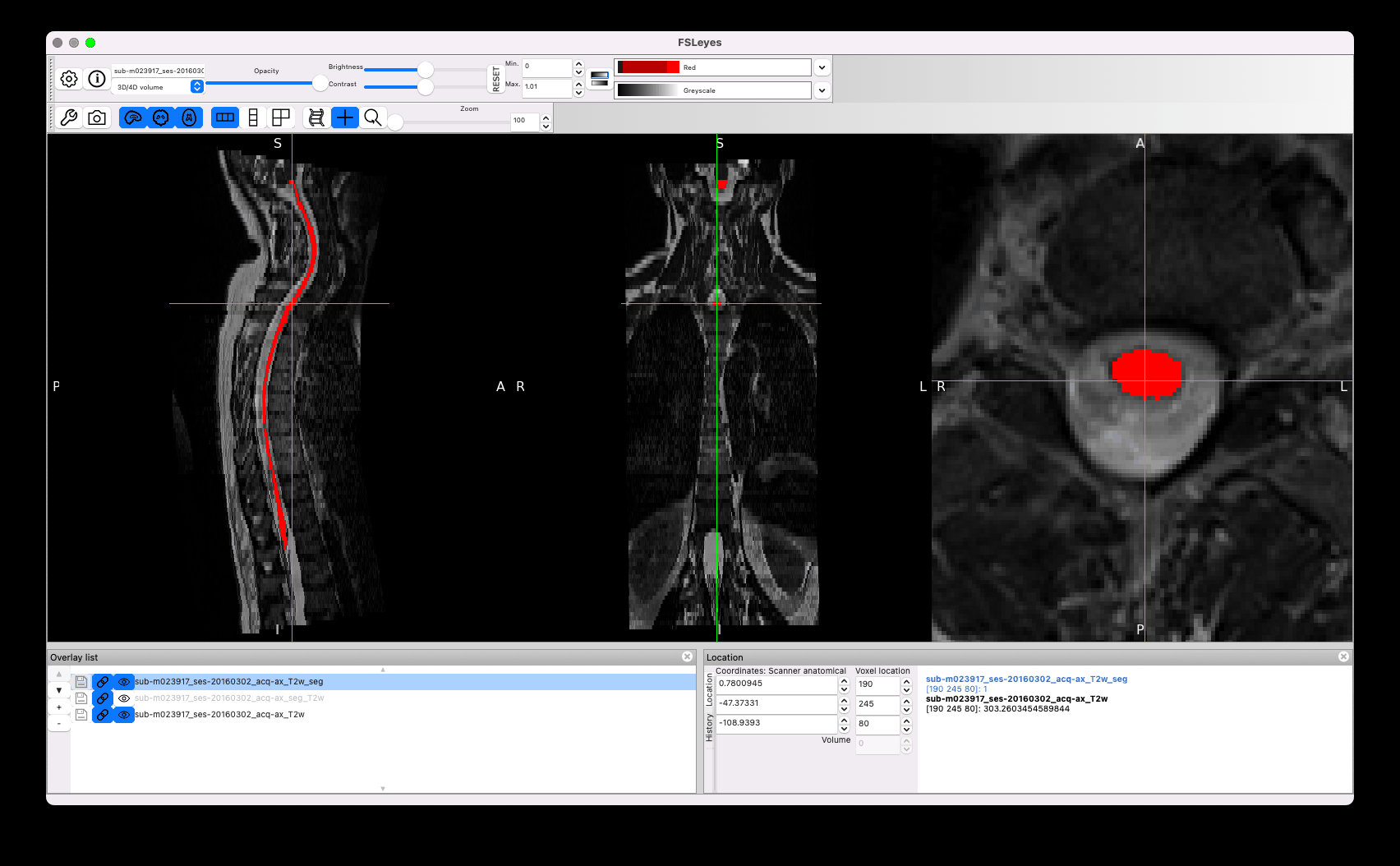
I am wondering why you did not start from sct_deepseg_sc, which seems to give overall better segmentation than the manual segmentation (red segmentation in the GIF above).
However, despite the better segmentation, the jittering still occurs:

2+ labels
Using 2+ labels implies a non-linear registration at step=0 to match the disc levels of the PAM50. Given the highly anisotropic resolution, maybe this non-linear registration is causing the jittering. I tried doing the registration to the template with only 2 discs:
sct_label_utils -i sub-m023917_ses-20160302_acq-ax_disks_T2w.nii.gz -keep 2,8 -o sub-m023917_ses-20160302_acq-ax_disks_T2w_2-8.nii.gz
sct_register_to_template -i sub-m023917_ses-20160302_acq-ax_T2w.nii.gz -s sub-m023917_ses-20160302_acq-ax_T2w_seg.nii.gz -ldisc sub-m023917_ses-20160302_acq-ax_disks_T2w_2-8.nii.gz -c t2 -r 0
However, the jittering is still there:
 IMPORTANT
IMPORTANT
Please be aware that when using 2+ labels, the image is cropped above/below the top/bottom labels. So make sure cropping does not happen in regions you are interested in, otherwise add more labels at the top/bottom, or use 2 labels only.
Straightening
Strangely, when running straightening outside of sct_register_to_template, there is no jittering, on both the manual and the deepseg-based segmentations:
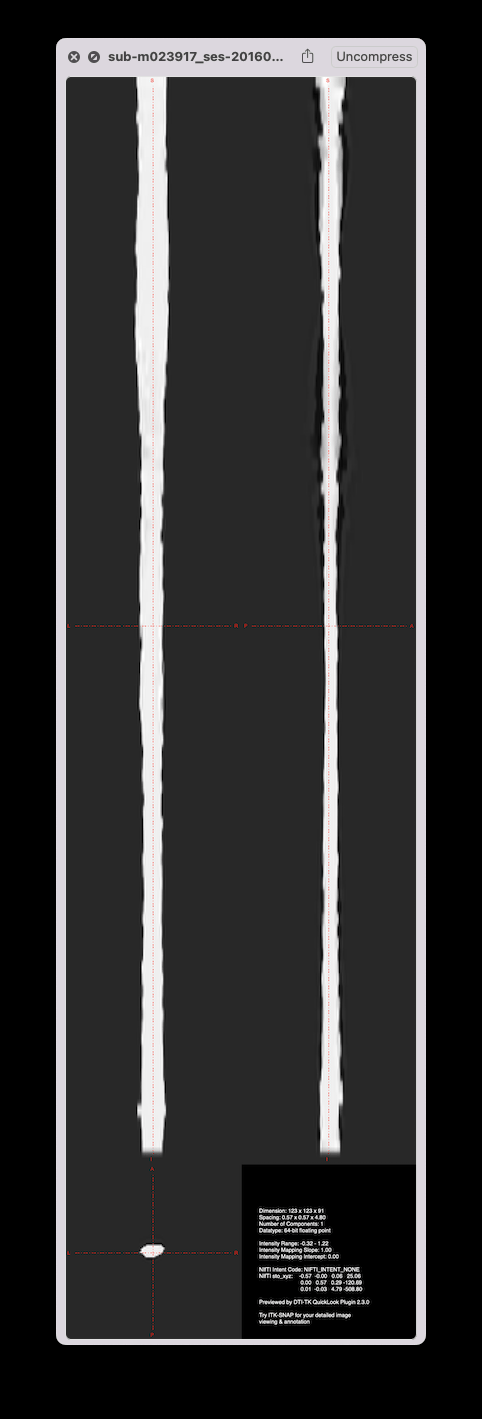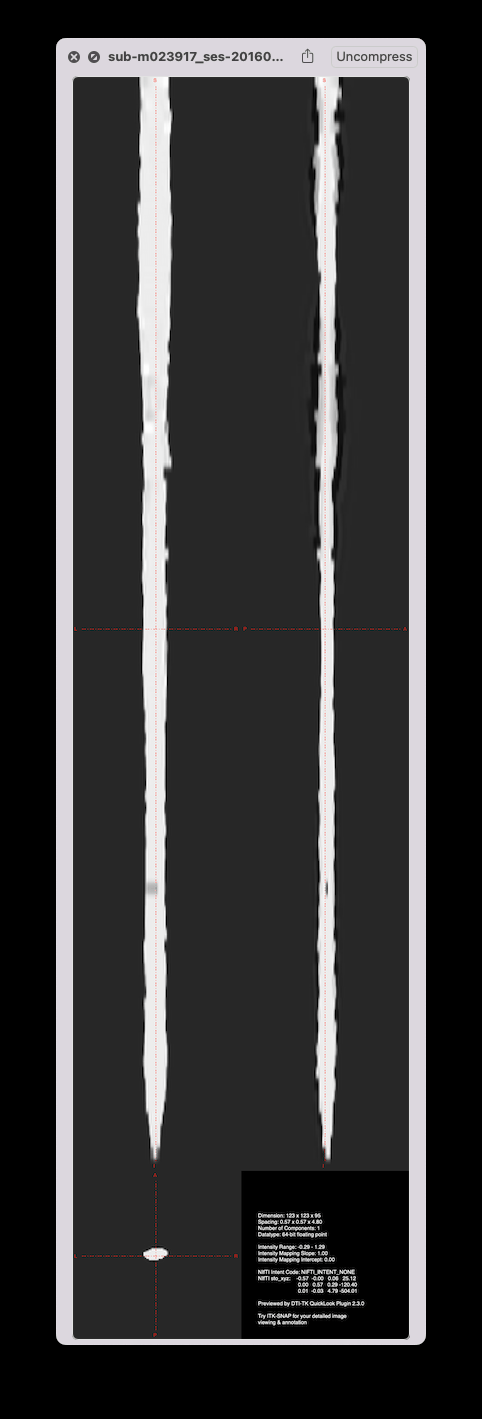
So, this suggests something fishy in sct_register_to_template pipeline. Digging deeper…
When looking at the straightening results on the intermediate outputs of the function (using -r 0 flag to access them), I notice that the straightening is performed on the 1mm resampled data. Now, one issue that was recently noticed, is that the resampling performed on binary masks with dtype UINT8 produces binary masks, even though linear interpolation is specified (for more details see issue: https://github.com/spinalcordtoolbox/spinalcordtoolbox/issues/4210). So let’s try to resample the segmentation properly to FLOAT32 with linear interpolation, and run the straightening:
sct_image -i seg_bin.nii.gz -type float32 -o seg_bin_float32.nii.gz
sct_resample -i seg_bin_float32.nii.gz -mm 1
sct_image -i seg_bin_float32_r.nii.gz -setorient RPI
sct_crop_image -i seg_bin_float32_r.nii.gz -zmin 148 -zmax 485
sct_straighten_spinalcord -i seg_bin_float32_r_crop.nii.gz -s seg_bin_float32_r_crop.nii.gz -dest template_seg_crop.nii.gz
Nope, that didn’t help:
However, when running the straightening on the native resolution, there is no jittering:
sct_straighten_spinalcord -i seg.nii.gz -s seg.nii.gz
This suggests that the issue is intrinsic to the native anisotropic resolution, and straightening images with axial images with large thickness will inevitably result in jittering, especially in presence of large curvature (ie: cases where the cord is not orthogonal to the slice).
Registration parameter
An obvious way to mitigate the jittering, is to regularize during registration. By default, sct_register_to_template uses centermassrot at step=1, so if there is any jittering, it will be passed on by centermassrot. Instead, I recommend using e.g. slicereg with high polynomial order (to accommodate complex cord curvatures) and relying on the image at step=2 for fine-tuning:
sct_register_to_template -i sub-m023917_ses-20160302_acq-ax_T2w.nii.gz -s sub-m023917_ses-20160302_acq-ax_T2w_seg.nii.gz -ldisc sub-m023917_ses-20160302_acq-ax_disks_T2w.nii.gz -c t2 -param step=1,type=seg,algo=slicereg,smooth=1,poly=5:step=2,type=im,algo=syn,metric=cc,iter=3
Results look nice in the native space:
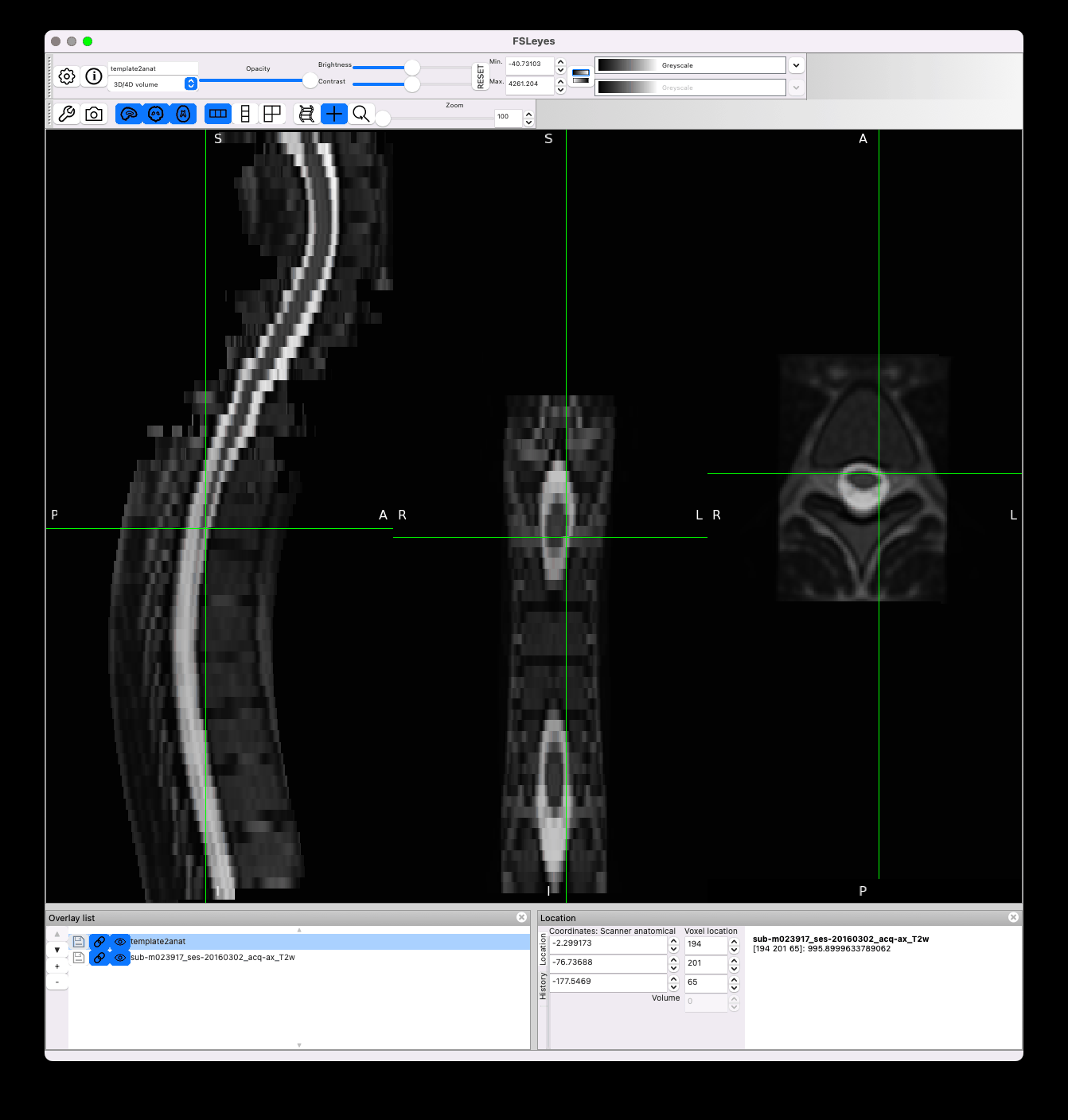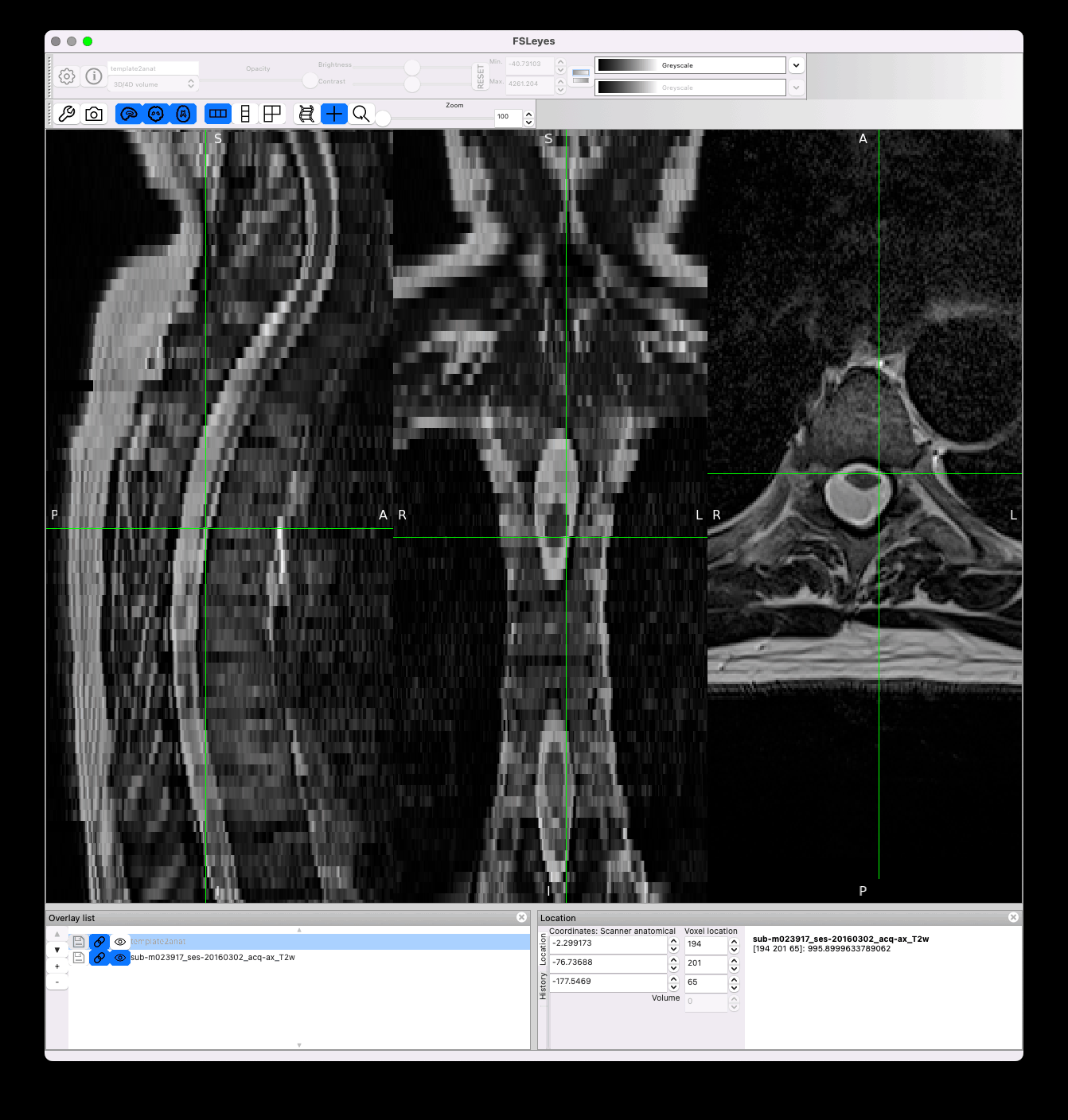
However there is still jittering in the template space, which I think is caused by the transformation of highly anisotropic resolution data coupled with high cord curvature:
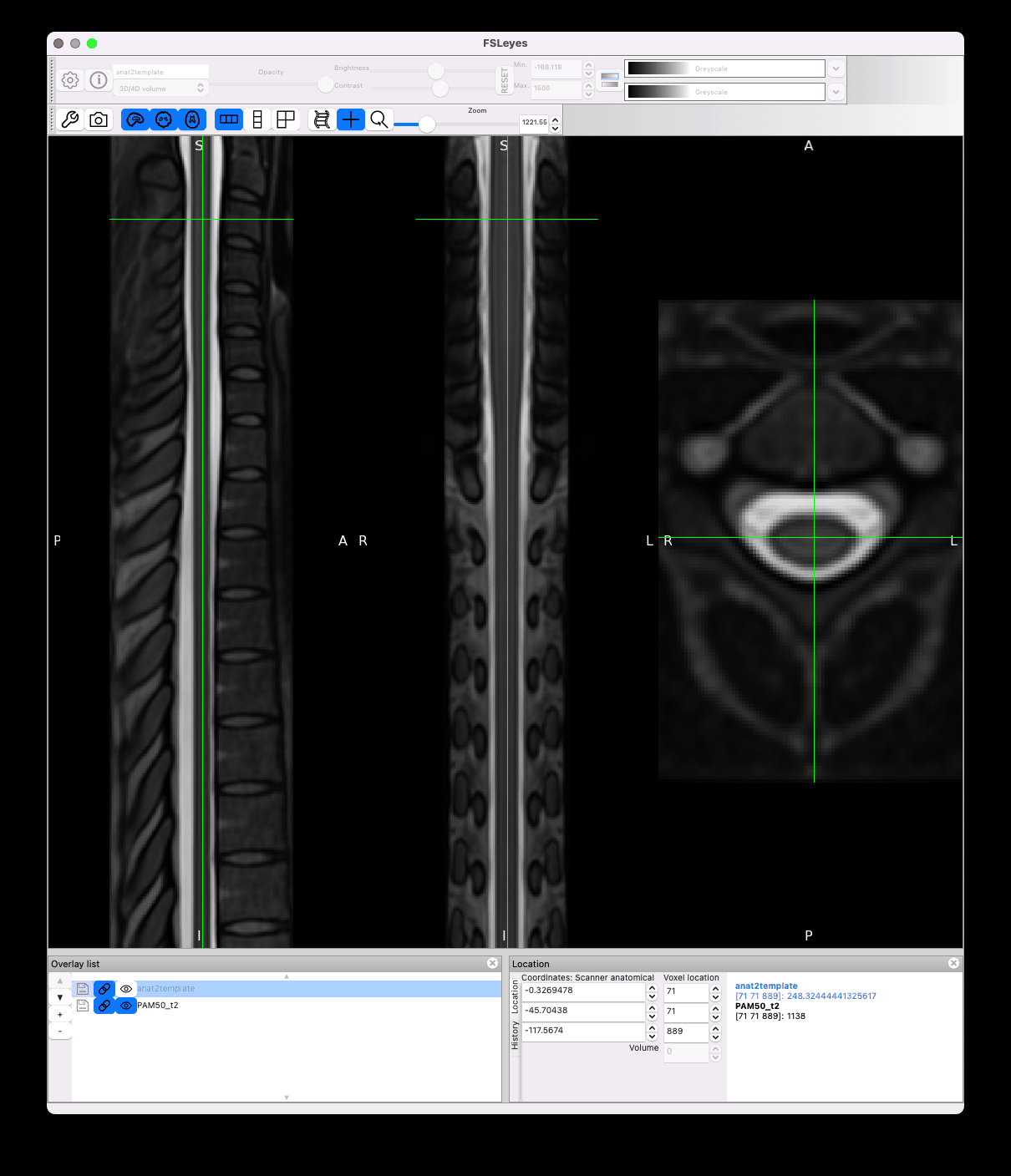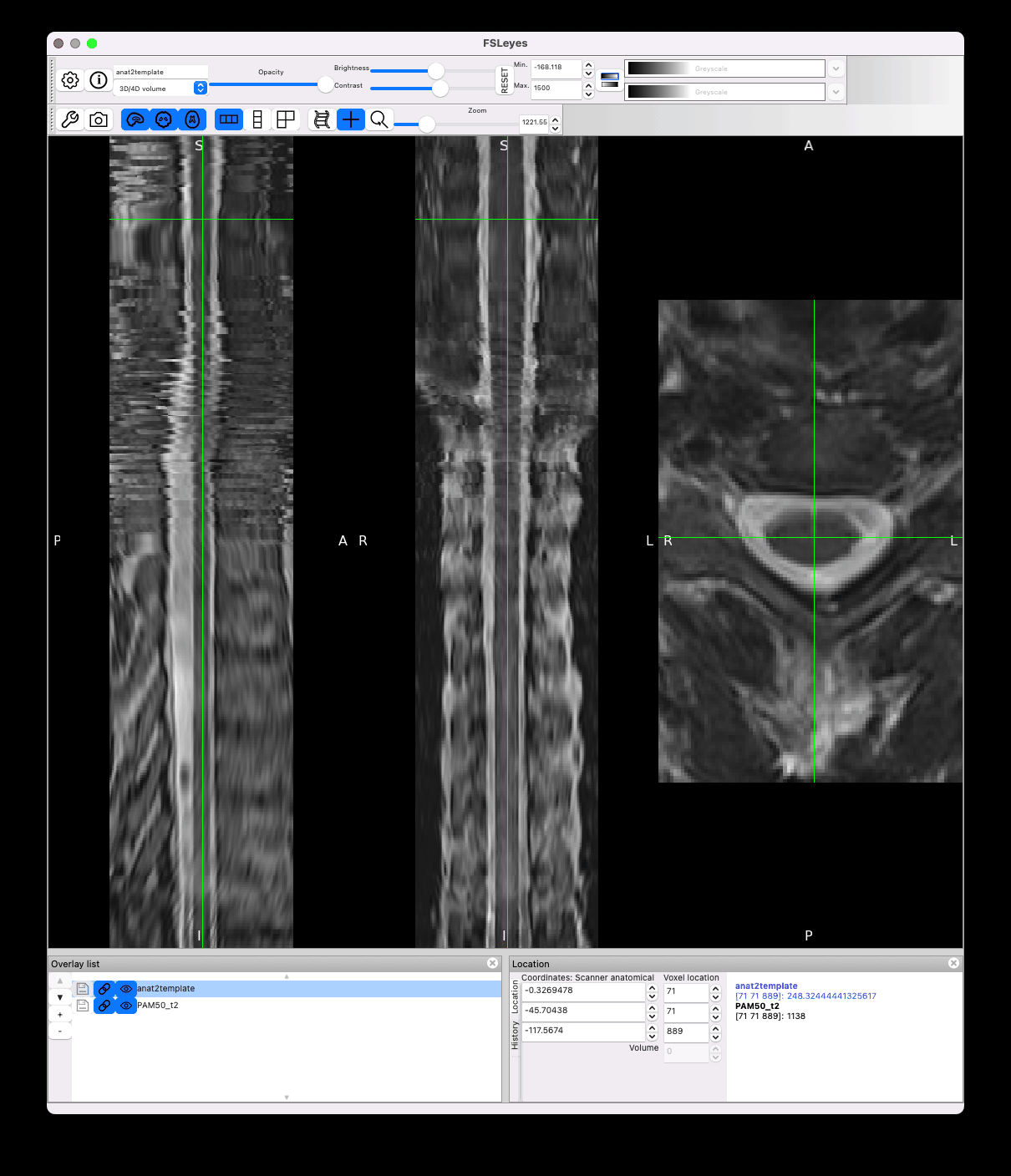
Unfortunately I don’t think there is anything we can do here, apart from adding smoothing on the input image, but that will reduce the effective resolution.
 IMPORTANT
IMPORTANT
I’ve only tried one set of registration parameter (not enough time). By spending some more time tweaking the registration parameters, you might be able to obtain even better results. Also, I’ve only tested on one subject, so make sure these params apply to your cohort.
Conclusion
My suggestions are:
- Improve the segmentation you are working with (use
sct_deepseg_sc and manually correct)
- Use the suggested registration parameters, which are possibly less sensitive to the jittering of the straightened segmentation at step=1,
- Do you really need to register to the PAM50 if your goal is to quantify lesions and CSA? Given the highly anisotropic nature of the data, I think you are better off doing the metric computation in the native space.
See my updated script with some comments: register_to_PAM50_manual_disks_cleaned_julien.sh (9.4 KB)