Hi Community
I’m trying to detect PMJ in a T2 image from the spine generic dataset, however i had this message
my command
sct_detect_pmj -i C:\Users\MSI\Desktop\sci\segsaye\sub-milan07_T2w_sc_seg.nii.gz -c t2 -qc ctrl
I’m working in Windows 10
Hi Community
I’m trying to detect PMJ in a T2 image from the spine generic dataset, however i had this message
my command
sct_detect_pmj -i C:\Users\MSI\Desktop\sci\segsaye\sub-milan07_T2w_sc_seg.nii.gz -c t2 -qc ctrl
I’m working in Windows 10

Hi,
I notice you input the segmentation instead of the actual image (the file after flag -i has the suffix _seg, so I assume it is a segmentation).
From the help:
sct_detect_pmj
--
Spinal Cord Toolbox (git-jca/4298-flow-d396ad3a9090900cc62d4b6eb4ccbc6d63d4a6eb)
sct_detect_pmj
--
usage: sct_detect_pmj -i <file> -c {t1,t2} [-h] [-s <file>] [-ofolder <folder>] [-o <file>]
[-qc <str>] [-qc-dataset <str>] [-qc-subject <str>] [-r {0,1}] [-v <int>]
Detection of the Ponto-Medullary Junction (PMJ). This method is based on a machine-learning
algorithm published in (Gros et al. 2018, Medical Image Analysis,
https://doi.org/10.1016/j.media.2017.12.001). Two models are available: one for T1w-like and another
for T2w-like images. If the PMJ is detected from the input image, a NIfTI mask is output
("*_pmj.nii.gz") with one voxel (value=50) located at the predicted PMJ position. If the PMJ is not
detected, nothing is output.
MANDATORY ARGUMENTS:
-i <file> Input image. Example: t2.nii.gz
-c {t1,t2} Type of image contrast, if your contrast is not in the available options (t1,
t2), use t1 (cord bright/ CSF dark) or t2 (cord dark / CSF bright)
Cheers,
Julien
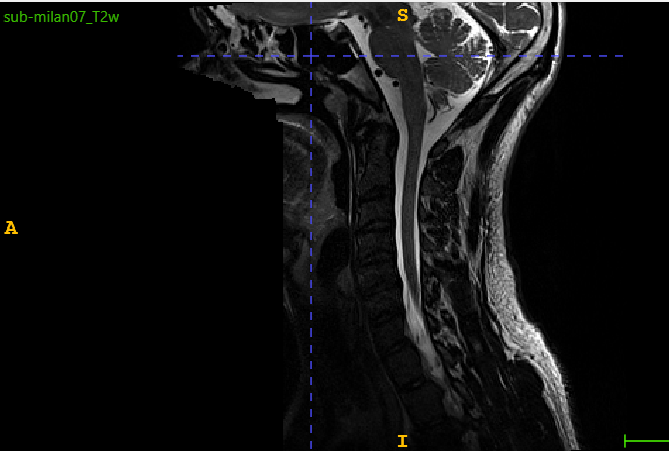
Yes, I see. Thank you for the remark, but I still have the same output
sct_detect_pmj -i C:\Users\MSI\Desktop\sci\segsaye\sub-milan07_T2w.nii.gz -c t2 -qc ctrl
Can you send me the image privately so I can try to reproduce this issue and understand its cause?
yes sure, i sent it to your email
I can reproduce your error. Unfortunately this is due to a sub-performance of the PMJ detection method-- it sometimes fails. I tried it on milan06 and it worked fine.
However I agree that the output message is not super informative, and this is something we can improve on our end @joshuacwnewton
cheers
Thanks for the ping! I’ll open up an issue on GitHub to follow up with a more intuitive error message. ![]()
Kind regards,
Joshua