Dear SCT Team:
"When I use the command ‘sct_deepseg_gm -i t2s.nii.gz’ to process 2D MRI data (t2s.nii), I obtain the result in t2s_gmseg.nii. What confuses me is that it seems like some slices are missing. Could you provide me with the reasons behind this issue and possible solutions?
t2.zip.001 (4 MB)
t2.zip.002 (729.4 KB)
Hi @Xujian_Wang,
I downloaded your data and tried running sct_deepseg_gm, and got the same results as you did (missing slices).
sct_deepseg_gm -i t2s.nii.gz
Taking a look at some of the empty slices from your t2s.nii.gz image:



And comparing them to the following image from one of SCT’s test dataset (which works well):

Comparing the two, my guess is that the issue is because of either (1) insufficient contrast between the gray and white matter of your scans and/or (2) increased resolution of the scans compared to the training data.
Still though, I tried various approaches to improve the quality of segmentation:
- Changing the
-moption ofsct_deepseg_gm - Changing the
-toption ofsct_deepseg_gm - Using the alterative GM segmentation approach:
sct_deepseg -task seg_gm_sc_7t_t2star - Cropping the image around the spinal cord.
I found that the last approach (cropping the image) worked well:
sct_deepseg_sc -i t2s.nii.gz -c t2s
sct_crop_image -i t2s.nii.gz -m t2s_seg.nii.gz -dilate 10
sct_deepseg_gm -i t2s_crop.nii.gz
Results:
I am curious as to why cropping the image has such a beneficial effect. (Especially because the test image from sct_example_data does not require cropping.) Maybe @jcohenadad can chime in?
Still though, I hope this helps. ![]()
Kind regards,
Joshua
Thank you for investigating, @joshuacwnewton. Indeed, the “missing slice” issue is caused by insufficient contrast between the gray and white matter of these scans.
First of all, thank you very much for the assistance you provided.
However, when I run the command “sct_crop_image -i t2s.nii.gz -m t2s_seg.nii.gz -dilate 10”, an error occurs as shown in the screenshot.
Can I use alternative commands instead of -dilate?
Thank you very much!
Hi @Xujian_Wang,
My apologies! The "-dilate" option was introduced in SCT v6.0 (#4020)
Here is an alternative that should work for older versions of SCT:
sct_deepseg_sc -i t2s.nii.gz -c t2s
# Dilate the spinal cord mask directly
sct_maths -i t2s_seg.nii.gz -dilate 10 -shape disk -dim 2 -o t2s_seg_dilate.nii.gz
# Pass the dilated mask to sct_crop_image
sct_crop_image -i t2s.nii.gz -m t2s_seg_dilate.nii.gz -o t2s_crop.nii.gz
sct_deepseg_gm -i t2s_crop.nii.gz
I double-checked, and it looks like this produces identical results to sct_crop_image -dilate 10.
Though, I would emphasize again that due to poor contrast, the results may not be as accurate as they could be with better contrast. So, I would take caution when using the segmentation produced here (making sure it matches your expectations, performing manual corrections where appropriate, etc.).
Kind regards,
Joshua
Thank you very much. I will process a subset of my data and observe the results to see if they meet expectations.
Hi @Xujian_Wang ,
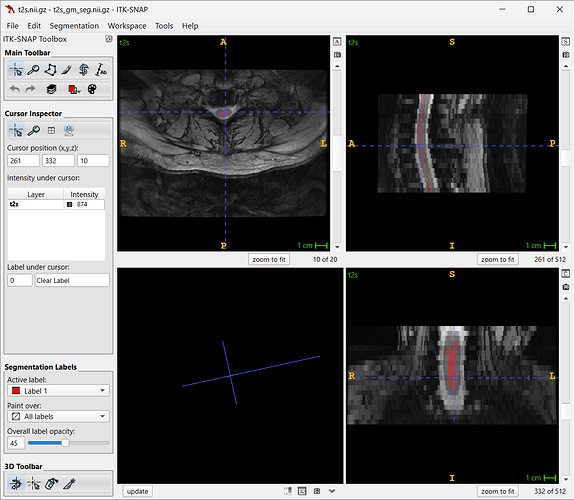
The SCTv.7.0 features the graymatter model, which is able to segment images offset to the center of matrix (which is why sct_deepseg_gm doesnot work well).
I have taken the liberty of testing it on your t2s.nii.gz image discussed here:
sct_deepseg graymatter -i t2s.nii.gz
Finding interesting results (no need to crop):
Best,
Nilser