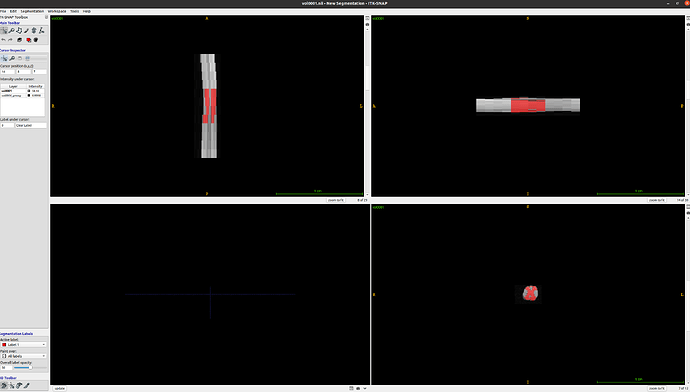
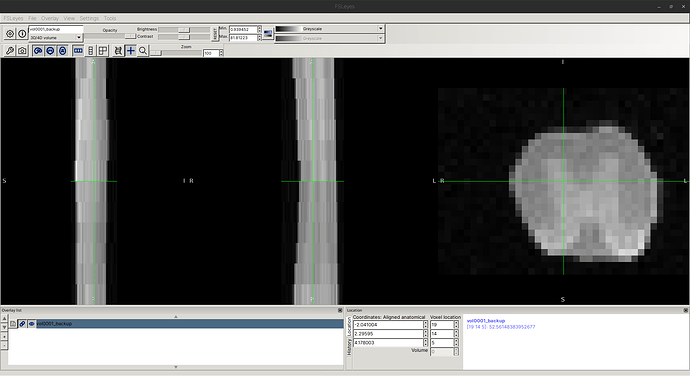
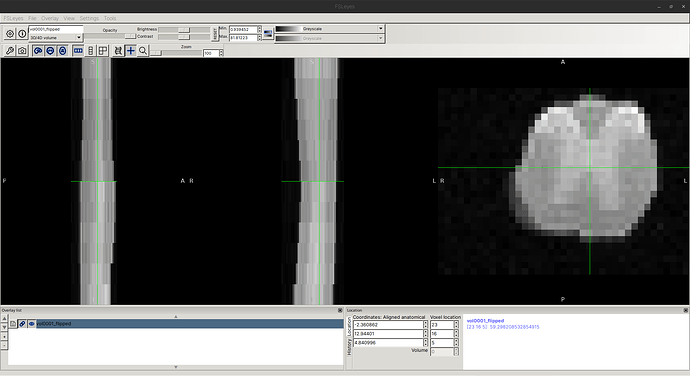
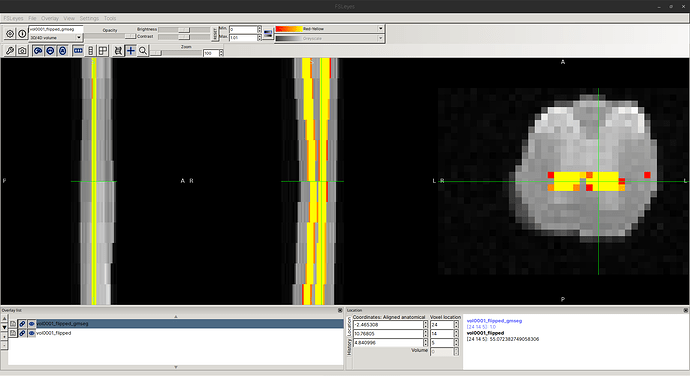
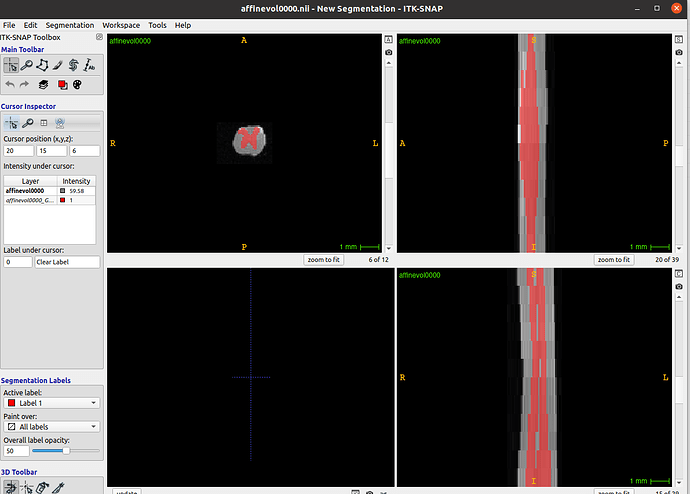
When I examine the header of the original image using sct_image -header, I see:
dim [3, 39, 29, 12, 1, 1, 1, 1]
pixdim [1.0, 0.078, 0.078, 1.0, 1.0, 1.0, 1.0, 1.0]
sform_name Aligned Anat
sform_code 2
sto_xyz:1 0.078000 0.000000 0.000000 -3.523000
sto_xyz:2 0.000000 0.000000 1.000000 -3.204000
sto_xyz:3 0.000000 -0.078000 0.000000 5.270000
sto_xyz:4 0.000000 0.000000 0.000000 1.000000
In other words, each axial slice is [39, 29] with [0.078, 0.078] mm resolution.
However, the actual affine is a bit strange: In the top-left 3x3 matrix, the zooms are not diagonal, as one might expect?
As a result, when I look at the internal state of sct_deepseg (after the images are loaded by our ivadomed deep learning package), I see:
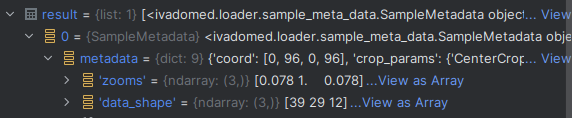
Suspiciously, the “zooms” do not match the axis order of the “data_shape”. I think this could be caused by the affine matrix, and I think this might explain the “empty slices” behavior (wrong zooms → wrong preprocessing → only part of the image is processed).
As a quick, hacky test, I tried to manually edit the affine (not recommended), and I was able to eliminate the “cropping” issues. So, I believe the affine is indeed the root cause of this behavior.
Before I continue further with manually editing the file, though, I want to stop and suggest that you look back to your image acquisition pipeline (DICOM → NIFTI conversion, etc.) and see if you have any steps that modify the affine/data array/header. Ideally, we would want to fix the data issue at the source, rather than performing these manual fixes to the data after the fact.
Kind regards,
Joshua