Dear Jcohenadad
I have a problem,
Firstly, the original file has 6 slices(0-5). but the csa of the FA only shows the slice 0-4. why this will happened.
Case108-1_DTI_thk7_breath-1_CLEAR_5_1.bval (63 Bytes) Case108-1_DTI_thk7_breath-1_CLEAR_5_1.bvec (832 Bytes) Case108-1_DTI_thk7_breath-1_CLEAR_5_1.nii.gz (658.3 KB)
These are my original file.
DWI_FA.csv (7.1 KB)
And this is my outcome.
dti_FA.nii.gz (53.0 KB)
This is the converted file of FA, it has only 5 slices (0-4).
What happened?
Secondly, when I deal with the imaging, sometimes it showed content below, then the value of slice 0,1,2,3 is empty. do you think I could continue to calculate the FA of such imaging? The derminal shows below.
Split destination segmentation…
Estimate cord angle for each slice: 0%| | 0/6 [00:00<?, ?iter/s]WARNING: Slice #0 is empty. It will be ignored.
WARNING: Slice #1 is empty. It will be ignored.
WARNING: Slice #2 is empty. It will be ignored.
Estimate cord angle for each slice: 100%|███████████████████████████| 6/6 [00:00<00:00, 77.49iter/s]
Build 3D deformation field: 100%|███████████████████████████████████| 3/3 [00:00<00:00, 89.51iter/s]
Best Wishes
Hi,
Firstly, the original file has 6 slices(0-5). but the csa of the FA only shows the slice 0-4. why this will happened.
What do you mean by CSA of the FA? What you calculated was not the CSA, but the computation of the FA within specified mask. Please clarify your question.
This is the converted file of FA, it has only 5 slices (0-4). What happened?
I downloaded your dti_FA.nii.gz and it has 6 slices:
$ fslhd dti_FA.nii.gz | grep dim3
dim3 6
Secondly, when I deal with the imaging, sometimes it showed content below, then the value of slice 0,1,2,3 is empty. do you think I could continue to calculate the FA of such imaging? The derminal shows below.
As the message says, the slices are empty. Check your segmentation, your registration and all slices of your DTI result.
Dear Jcohenadad
I am sorry, I uploaded a wrong DTI file, this is the corresponding file to the original file.
This is my original file.
Case097-1_DTI_thk7_breath-1_CLEAR_5_1.PAR (34.6 KB) Case097-1_DTI_thk7_breath-1_CLEAR_5_1.REC (3.2 MB)
Case097-1_DTI_thk7_breath-3_CLEAR_5_3.nii.gz (605.2 KB)
this is the converted file.
dti_FA.nii.gz (34.1 KB)
This one has only 4 slices.
Why will it happen?
Secondly, I used the same script to run the different files (about 150), However some files (about 20) could not get the result, for example this file.
Case086-1_DTI_thk7_breath-1_CLEAR_7_1.PAR (34.6 KB) Case086-1_DTI_thk7_breath-1_CLEAR_7_1.REC (3.2 MB)
Case086-1_DTI_thk7_breath-1_CLEAR_7_1.bval (67 Bytes) Case086-1_DTI_thk7_breath-1_CLEAR_7_1.bvec (836 Bytes) Case086-1_DTI_thk7_breath-1_CLEAR_7_1.nii.gz (674.2 KB)
and the terminal shows: Command-line usage error: Option -i file dti_FA.nii.gz does not exist.
Aborted…
and this is my script.
861.sh (3.0 KB)
why some files show this errors, the others will not?
Best Wishes
This is my original file.
Case097-1_DTI_thk7_breath-1_CLEAR_5_1.PAR (34.6 KB) Case097-1_DTI_thk7_breath-1_CLEAR_5_1.REC (3.2 MB)
Case097-1_DTI_thk7_breath-3_CLEAR_5_3.nii.gz (605.2 KB)
this is the converted file.
dti_FA.nii.gz (34.1 KB)
This one has only 4 slices.
Why will it happen?
Hi, I’m sorry but this forum is dedicated to SCT issues only. If you have problems with Dicom conversion, I suggest you talk to your local IT expert.
Secondly, I used the same script to run the different files (about 150), However some files (about 20) could not get the result, for example this file.
Case086-1_DTI_thk7_breath-1_CLEAR_7_1.PAR (34.6 KB) Case086-1_DTI_thk7_breath-1_CLEAR_7_1.REC (3.2 MB)
Case086-1_DTI_thk7_breath-1_CLEAR_7_1.bval (67 Bytes) Case086-1_DTI_thk7_breath-1_CLEAR_7_1.bvec (836 Bytes) Case086-1_DTI_thk7_breath-1_CLEAR_7_1.nii.gz (674.2 KB)
and the terminal shows: Command-line usage error: Option -i file dti_FA.nii.gz does not exist.
Aborted…
When this happens, scroll up in your Terminal window and check which previous process failed (you likely got other errors that led to the dti_FA.nii.gz file not being created). Once you’ve identified the first process that failed, copy/paste the command that failed and send the relevant files. Then, I will be able to help.
Dear Jcohenadad
For the first problem, there is no problem with Dicom conversion, the converted file of nii.gz still has 6 slices, Case097-1_DTI_thk7_breath-3_CLEAR_5_3.nii.gz (605.2 KB)
however when the Terminal runs the script, the DTI file became 5 slices.dti_FA.nii.gz (34.1 KB)
What goes wrong?
Secondly, I think maybe the process failed here
Apply transformation and resample to destination space…
/Users/hkudkch/sct_4.2.1/bin/isct_antsApplyTransforms -d 3 -i src_seg.nii -o src_seg_reg.nii -t warp_forward_0.txt -r dest_seg_RPI.nii -n NearestNeighbor # in /private/var/folders/t_/7lybdzgj0fz4wp9zpdg19cnr0000gn/T/sct-20200223190617.042421-register-8bbu34zr
Copy affine matrix from destination space to make sure qform/sform are the same.
WARNING: the resulting image could have wrong apparent results. You should use an affine transformation as last transformation…
But many files run normally still show this line.
Or the. Command-line usage error: Option -i file dti_FA.nii.gz does not exist.
Aborted…
The Terminal only show this wrong warning.
This is the file.
Case086-1_DTI_thk7_breath-1_CLEAR_7_1.bval (67 Bytes) Case086-1_DTI_thk7_breath-1_CLEAR_7_1.bvec (836 Bytes) Case086-1_DTI_thk7_breath-1_CLEAR_7_1.nii.gz (674.2 KB)
Case086-1_DTI_thk7_breath-1_CLEAR_7_1.PAR (34.6 KB) Case086-1_DTI_thk7_breath-1_CLEAR_7_1.REC (3.2 MB)
Best Wishes
For the first problem, there is no problem with Dicom conversion, the converted file of nii.gz still has 6 slices, Case097-1_DTI_thk7_breath-3_CLEAR_5_3.nii.gz (605.2 KB)
however when the Terminal runs the script, the DTI file became 5 slices.dti_FA.nii.gz (34.1 KB)
What goes wrong?
I have downloaded your file and ran this script 861.sh (3.0 KB), and the resulting file dti_FA.nii.gz has 6 (not 5) slices:
fslhd dti_FA.nii.gz | grep dim3
dim3 6
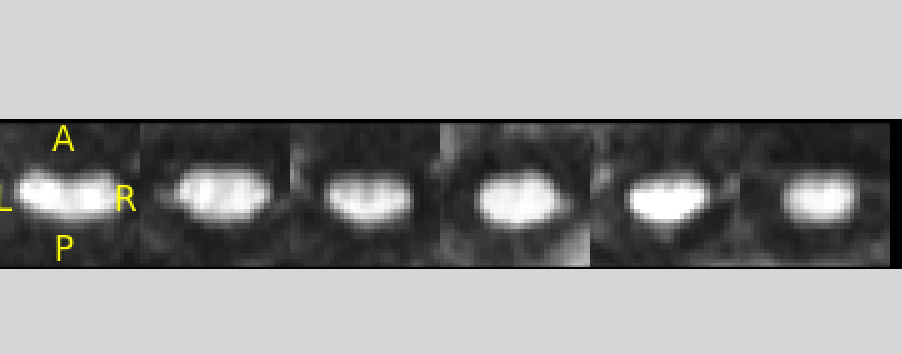
You should also be aware that the registration result is pretty bad, so you should adjust the registration parameters (see SCT course). See QC report here: qc.zip (378.3 KB)
Secondly, I think maybe the process failed here
Apply transformation and resample to destination space…
/Users/hkudkch/sct_4.2.1/bin/isct_antsApplyTransforms -d 3 -i src_seg.nii -o src_seg_reg.nii -t warp_forward_0.txt -r dest_seg_RPI.nii -n NearestNeighbor # in /private/var/folders/t_/7lybdzgj0fz4wp9zpdg19cnr0000gn/T/sct-20200223190617.042421-register-8bbu34zr
Copy affine matrix from destination space to make sure qform/sform are the same.
WARNING: the resulting image could have wrong apparent results. You should use an affine transformation as last transformation…
But many files run normally still show this line.
Or the. Command-line usage error: Option -i file dti_FA.nii.gz does not exist.
Aborted…
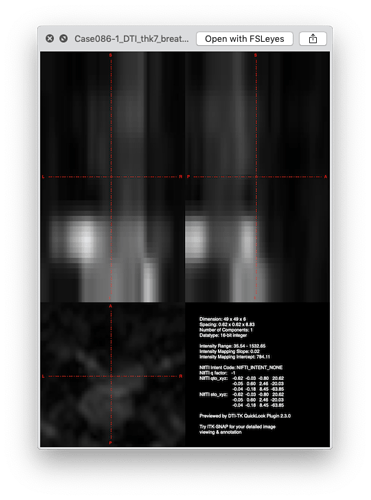
The image is corrupted, see below:
So in conclusion, I recommend you make sure to QA your images (i.e., open a viewer and look at your images) before running any processing on them.
Dear Jcohenadad
Thank you for your reply, do you think the registration result will effect the FA value of the each slice a lot?
sct_register_to_template -i ${file_dwi_mean}.nii.gz -s ${file_dwi_seg}.nii.gz -l label_C3.nii.gz -ref subject -c t1 -param step=1,type=seg,algo=centermassrot:step=2,type=im,algo=syn,metric=CC,slicewise=0,smooth=0,iter=6 -qc qc
mv warp_template2anat.nii.gz warp_template2dwi.nii.gz
mv warp_anat2template.nii.gz warp_dwi2template.nii.gz
and this is my registration parameters
I just wan to get the FA value of each slice, do you think where is the problem in this registration parameters.
I think I could not find the errors in this command, could you figure it out for me?
Maybe it is related to the sct_extract_metric?
sct_extract_metric -i dti_FA.nii.gz -f label/atlas -l 30,31,32,33,34,35,58,59 -perslice 1 -o DWI_FA.csv -append 1
This is my extract command line.
Do you mean this qc result is very bad?
I am a little confused.
Best Wishes
Dear @DKCH_HKU,
With all due respect, all these questions are already answered in my previous posts or in the SCT course. As my time is very limited, I can only answer questions that are “high level” and do not have the time to dive too much into the basics of image analysis. I think that you would need a more extensive and personalized help to perform the analysis for your research.
That being said, I would be happy to help and assist you with your analysis, but first we would need to officialize a collaboration. Please feel free to contact me via email if you are interested in such collaboration.
Best,
Julien
Hi,
So, I’ve looked at subject Case097-1_DTI_thk7_breath-3_CLEAR_5_3 and because of the large deformation, I suggest you use the algorithm centermassrot:
sct_register_to_template -i ${file_dwi_mean}.nii.gz -s ${file_dwi_seg}.nii.gz -l label_C2.nii.gz -ref subject -c t1 -param step=1,type=seg,algo=centermassrot:step=2,type=seg,algo=columnwise -qc qc
Here is the full script + the data: Case097.zip (603.0 KB)
Best,
Julien