Hi,
Yes and no. There is the function sct_maths -erode, but it uses a 3D approach. Coincidentally I am currently working on updating the function to allow 2D kernel. More info here. Stay tuned ![]()
Hi
I have tried to figure it out all the issues I had when dealing with neonatal DKI images. My aim was calculating average DKI and DTI maps over lateral CST tracts and later over other WM tracts. I had issues both in segmenting the cord and registering PAM50 atlas to neonatal subject. Per each subject, I have DKI and 3dT1 images. In the beginning I thought of using the 3dT1 image as a bridge (better anatomical reference) for registration of atlas to subject. However, sct_register_to_template from 3dT1 to PAM50 and viceversa produced an error , as already mentioned in previous discussions on the forum. So, I opted for using 3dT1 images just for label creations with interactive viewer, since discs are not well visible on DKI image.
As regards registration to the template, the problem I had was that the registration output I obtained included some CSF at some SC level due to size differences between PAM 50 ADULT atlas and my NEONATE images. So, in order to be sure to exclude CSF, I reduced PAM50 labels corresponding to the WM tract of interest (CST in this case) to just their intersection with SC segmentation of my images.
I used sct to compute DTI maps and DIPY to compute DKI maps.
Here you find attached the script with the processing steps (script_kurtosis.sh),the python DIPY script (dipy_DKI.py), all the images produced through applying the script, the DTI and DKI maps, and the average metrics results over lateral CST.
Do you think this is the resolutive approach? Could it work with other WM tracts?
I cannot clearly see if registration of the atlas to the template is fine also longitudinally, with a perfect correspondence along spinal levels, due to different sizes.
Thanks a lot,
Rosella
https://drive.google.com/drive/folders/1kaaz1_zuTsK7dPL4yuR1q4Agm8sH7p17
Hi Rosella,
However, sct_register_to_template from 3dT1 to PAM50 and viceversa produced an error , as already mentioned in previous discussions on the forum.
I see you had issues with sct_label_vertebrae here, but I cannot find the specific issue you encountered with sct_register_to_template. Which issue are you referring to?
As regards registration to the template, the problem I had was that the registration output I obtained included some CSF at some SC level due to size differences between PAM 50 ADULT atlas and my NEONATE images. So, in order to be sure to exclude CSF, I reduced PAM50 labels corresponding to the WM tract of interest (CST in this case) to just their intersection with SC segmentation of my images.
I used sct to compute DTI maps and DIPY to compute DKI maps.
Here you find attached the script with the processing steps ( script_kurtosis.sh ),the python DIPY script ( dipy_DKI.py ), all the images produced through applying the script, the DTI and DKI maps, and the average metrics results over lateral CST.
Do you think this is the resolutive approach? Could it work with other WM tracts?
A couple of comments:
- Why are you outputting the CSF from the propseg segmentation?
- Your script has an error at line 12:
-ofolderis missing an argument. - No, it is not a good idea to reduce the intersection between the registered PAM50 data (e.g. WM) and your cord segmentation, because (i) if there is a misregistration, simply taking the intersect will not solve the other issues (e.g. misregistration inside the cord) and (ii) modifying the partial volume information will violate the assumption made while running
sct_extract_metricusing maximum likelihood or maximum a posteriori estimation. The right thing to do is change the registration parameter to allow more degree of freedom, which is what i did in the updated script. - The data you shared in your previous post do not include the file
3dT1.nii, therefore I cannot get the labels from this file. So instead, I assumed realistic vertebral levels for two slices on the kurtosis image directly, and I used that to generate two labels. You will need to update those numbers according to the proper levels. - I recommend you use the QC feature. It help validating the quality of segmentation, registration to template, etc. and also to share your analysis and keep that as a record of what you actually ran (because the exact syntax is included in the QC report, along with SCT version).
So, I modified the script to address all those issues: kurtosis_JCA.sh (2.7 KB) . Here are the QC report and DTI results so you can make sure you can reproduce my results: qc-dti.zip (428.6 KB)
I cannot clearly see if registration of the atlas to the template is fine also longitudinally, with a perfect correspondence along spinal levels, due to different sizes.
Please note that your workflow is using vertebral levels as references for alignment with the PAM50 template, not spinal levels.
Best,
Julien
I refer to quality of registration, which was not good between 3dT1 and PAM50 atlas.
I wanted to check if part of my SC segmentation took also a part of CSF.
I didn’t understand this step.
Yes, I used an improper term, sorry!
Anyway, do you think registering the PAM50 atlas directly to DKI image is the right approach?
Should I try again to use 3dT1 image as a bridge for registration ?
Finally, do you think through your script registration is good enough for ROI definition or is manual ROI definition preferred?
Thank you very much,
Rosella
here is my 3dT1 image
https://drive.google.com/drive/u/0/folders/1kaaz1_zuTsK7dPL4yuR1q4Agm8sH7p17
best,
Rosella
Moreover, which unit measures does sct_dmri_compute_dti use?
thanks,
Rosella
Hi,
one more thing . When I compute DKI metrics using DIPY in the python script, do I have to use the dilated mask all the same ?
Thanks,
Rosella
I didn’t understand this step.
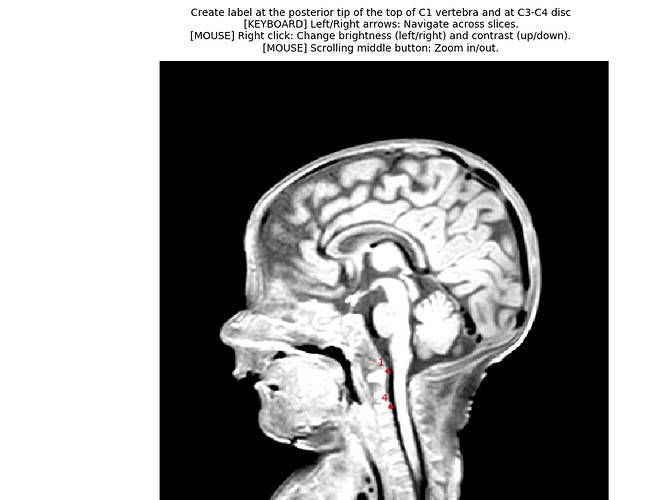
What I meant is that I opened the mean DWI image and I tried to look for the visible intervertebral discs, to find what slice number corresponds to disc 1 and C3/C4. But if you have access to your 3D T1 image, then you can use that for more accuracy. Please note that I’ve changed the script to label 1 and 4 (instead of 1 and 3). Here is the updated script: kurtosis_JCA_20200302103420.sh (2.8 KB)
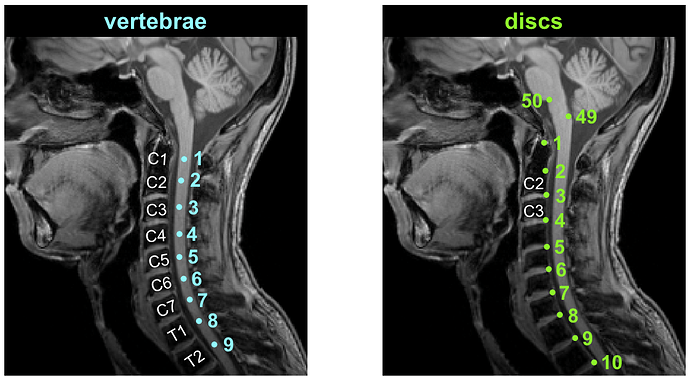
For disc label definition, please see: SCT course page 47, also shown below for convenience:
Anyway, do you think registering the PAM50 atlas directly to DKI image is the right approach?
yes
Should I try again to use 3dT1 image as a bridge for registration ?
no-- you would accumulate errors, because the PAM50 → T1w is not reliable in your case (very small structure, different contrasts compared to adult, etc.)
Finally, do you think through your script registration is good enough for ROI definition or is manual ROI definition preferred?
I recommend using my script. Not only is it more accurate, but it is also less user biased and makes a reproducible pipeline.
Moreover, which unit measures does sct_dmri_compute_dti use?
sct_dmri_compute_dti is built on Dipy, so it is using the same units as Dipy (and the majority of other DW software). E.g., bmatrix is in s/mm^2.
When I compute DKI metrics using DIPY in the python script, do I have to use the dilated mask all the same ?
Use exactly the same as what is in the script I sent you (you can just add the DKI metrics inside the for loop)
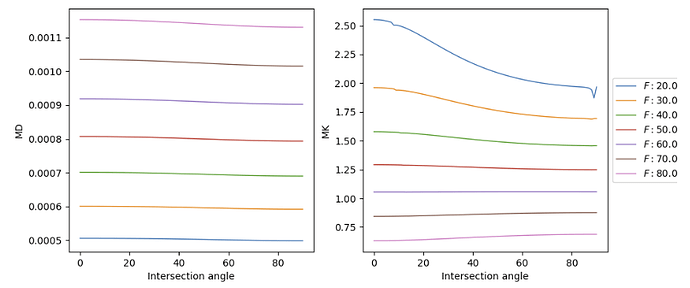
I am asking this because DTI metrics values appear to be very low, whereas DKI values seem more ‘normal’
Here you find attached results for MD and MK.
thanks,
RosellaDWI_MK.csv (448 Bytes) DWI_MD.csv (467 Bytes)
Is this label choice appropriate? I do not trust my self in marking labels
I am not an expert in pediatric spine.
I am asking this because DTI metrics values appear to be very low, whereas DKI values seem more ‘normal’. Here you find attached results for MD and MK.
MD and MK are not supposed to be “similar”. See below representative values (from: dipy website):
I can see that …I just wanted to know if the two labels required in your script could be ok in the screenshot I attached.
ok…so this agrees with my results.
Thank you,
Rosella
Hi,
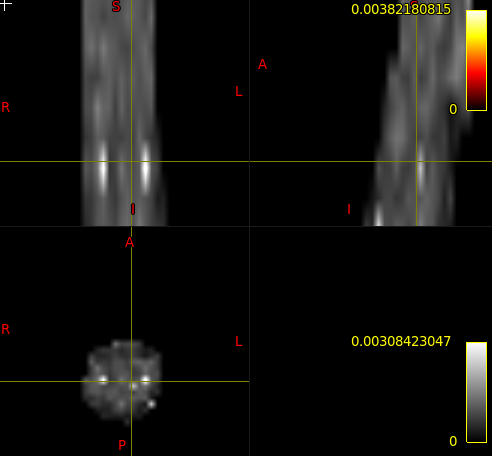
I was wondering what these white spots on DTI maps computed with SCT are due to.
Is there a way to remove them if artefactual?
Thanks,
Rosella
One more thing I have read sct_dmri_compute_dti relies on Dipy for DTI maps computation. Does this command use DTI tensor to compute DTI metrics or higher order DKI tensor for better accuracy?
Thanks,
Rosella
The same problems of this time black spots appears on DKI maps computed with DIPy
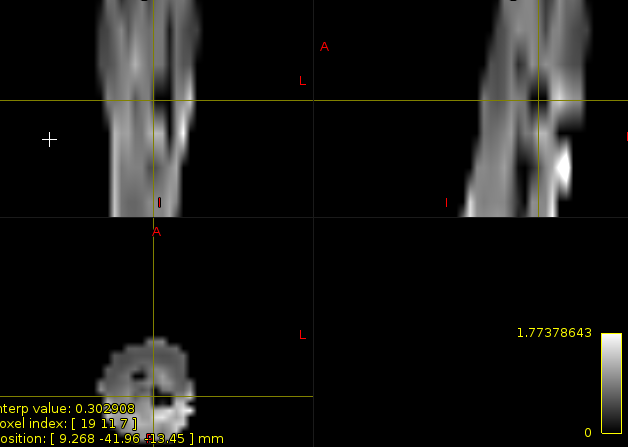
Do these artifacts influence metrics’ extraction and computation?
best,
Rosella
Hi Rosella,
I was wondering what these white spots on DTI maps computed with SCT are due to.
Is there a way to remove them if artefactual?
[…]
The same problems of this time black spots appears on DKI maps computed with DIPy
It could be noise in the DWI data or a genuine bright signal if the underlying structure reflects it (e.g. highly fibrous structure will produce high FA). I would suggest to not call them “DTI maps”. DTI is a mathematical model, from which you can derive metrics such as FA, MD, etc. The map you are showing is one of those metric.
Do these artifacts influence metrics’ extraction and computation?
Yes, so you might want to mask them out, if it is well justified.
One more thing I have read sct_dmri_compute_dti relies on Dipy for DTI maps computation. Does this command use DTI tensor to compute DTI metrics or higher order DKI tensor for better accuracy?
It is using the diffusion tensor model. Please note that while DK model might provide “better accuracy”, it is also hampered by lower precision (the more parameters in a model, the noisier the output metrics).
OK thank you very much for your answer. I will decide whether masking those spots.
Moreover, I am trying the segmentation and processing script on another subject. I have decided to keep the cropping as the first subject (C1-C4) for coherence. However, segmentation with deepseg seems giving worst results I think. Could it be ok to process this subject differently?Of course I’d rather find a processing suitable for all subjects.
Here you find attached the images
https://onedrive.live.com/?id=B0C0561E34E9D76F!818&cid=B0C0561E34E9D76F
hi, i cannot get the file (it says “item no longer exists”)
Hi. I have sent you an email with the sharing link.
Thanks,
Rosella
Moreover,
I cannot understand how you decided to crop the image in this case:
sct_crop_image -i kurtosis.nii -xmin 60 -xmax 100 -ymin 60 -ymax 100 -zmin 8 -o kurtosis_crop.nii
Especially, how can I be sure to crop all subjects the same way?
Thanks,
Rosella