I need to segment DKI neonatal images in order to compute DTI and DKI metrics over certain WM tracts. Segmentation is the first step of the pipeline.
I have tried with the following command:
sct_dmri_separate_b0_and_dwi -i kurtosis.nii -bvec bvecs -ofolder ./
sct_propseg -i kurtosis_dwi_mean.nii -c t2 -ofolder ./ -init-mask viewer
However, neither giving an initial mask or centerline does not give good results. I have also tried smoothing the image before segmentation or using the -rescale option with scale factor 2 without improved results.
here is a link to my data.
https://onedrive.live.com/?id=B0C0561E34E9D76F!596&cid=B0C0561E34E9D76F
Hi,
The link to the data does not work (it says “this item is no more available”), so i used the data you shared with me via email. There are 3 folders: subject_1, subject_2 and subject_3.
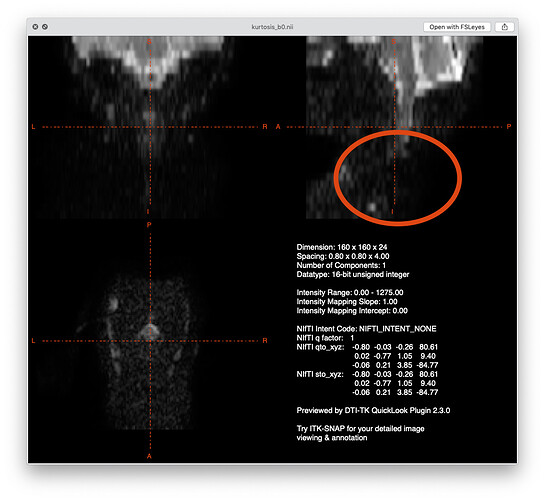
When looking at the data from subject_1, I can immediately see that half of the FOV is not usable due to poor image quality:
For the upper part of the cervical cord, I remember we already had an exchange about segmenting the cord and I believe I’ve already addressed your question here.
Best
Julien
yes, the lower part of the image is not usable so I decided to focus on upper part. That segmentation pipeline gets better results but does take some CSF and that is what I want to avoid.
best,
Rosella
Unfortunately with this image quality there is no reliable way to segment the cord in a perfect manner, without any manual intervention. So, I would suggest you do a first pass of deepseg_sc (or propseg, whichever works best for you) and then do a manual correction.
The next problem I have is in creating label vertebrae for 3dT1 image in order to register PAM50 atlas to my 3dT1 image and define ROIs over the registered atlas.
sct_label_vertebrae -i 3dT1.nii -s 3dT1_seg.nii -c t1 -ofolder ./
--
Spinal Cord Toolbox (4.2.1)
Folder ./ has been created.
Create temporary folder (/tmp/sct-20200128160925.047520-label_vertebrae-sny99fmr)...
Copying input data to tmp folder...
Straighten spinal cord...
Create temporary folder (/tmp/sct-20200128161200.307226-straighten_spinalcord-98rbu9s7)...
Copy files to tmp folder...
Window length needs to be >= 3. Returning input signal.
Window length needs to be >= 3. Returning input signal.
Fitting centerline using B-spline approximation
Error on approximation = 0.99 mm
Error on approximation = 0.36 mm
Error on approximation = 0.38 mm
Error on approximation = 0.16 mm
Error on approximation = 0.09 mm
Error on approximation = 0.06 mm
Error on approximation = 0.06 mm
Error on approximation = 0.06 mm
Error on approximation = 0.03 mm
Error on approximation = 0.04 mm
Error on approximation = 0.03 mm
Error on approximation = 0.02 mm
Error on approximation = 0.02 mm
Error on approximation = 0.02 mm
Error on approximation = 0.02 mm
Create the straight space and the safe zone
Length of spinal cord: 63.52542526347554
Size of spinal cord in z direction: 56.998004713880285
Ratio length/size: 1.1145201587733067
Safe zone boundaries (curved space): [-63.89699063260217, -6.898985918721884]
Safe zone boundaries (straight space): [-67.16070090739979, -3.635275643924256]
Pad input volume to account for spinal cord length...
Time to generate centerline: 52.0 ms
100%|█████████████████████████████████████████| 161/161 [01:06<00:00, 2.44it/s]
100%|█████████████████████████████████████████| 538/538 [10:35<00:00, 1.18s/it]
Warping field generated: tmp.curve2straight.nii.gz
Warping field generated: tmp.straight2curve.nii.gz
Apply transformation to input image...
/home/rosella/sct_4.2.1/bin/isct_antsApplyTransforms -d 3 -r tmp.centerline_pad_crop.nii.gz -i data.nii -o tmp.anat_rigid_warp.nii.gz -t tmp.curve2straight.nii.gz -n 'BSpline[3]' # in /tmp/sct-20200128161200.307226-straighten_spinalcord-98rbu9s7
Generate output files...
File created: ./warp_curve2straight.nii.gz
File created: ./warp_straight2curve.nii.gz
cp /tmp/sct-20200128161200.307226-straighten_spinalcord-98rbu9s7/tmp.anat_rigid_warp.nii.gz ./straight_ref.nii.gz
sct_convert -i /tmp/sct-20200128161200.307226-straighten_spinalcord-98rbu9s7/tmp.anat_rigid_warp.nii.gz -o ./data_straight.nii
File created: ./data_straight.nii
Remove temporary files...
rm -rf /tmp/sct-20200128161200.307226-straighten_spinalcord-98rbu9s7
Finished! Elapsed time: 1352 s
Done! To view results, type:
fsleyes ./data_straight.nii &
Resample to 0.5mm isotropic...
sct_resample -i data_straight.nii -mm 0.5x0.5x0.5 -x linear -o data_straightr.nii # in /tmp/sct-20200128160925.047520-label_vertebrae-sny99fmr
Apply straightening to segmentation...
/home/rosella/sct_4.2.1/bin/isct_antsApplyTransforms -d 3 -i segmentation.nii -r data_straightr.nii -t warp_curve2straight.nii.gz -o segmentation_straight.nii -n Linear # in /tmp/sct-20200128160925.047520-label_vertebrae-sny99fmr
sct_maths -i segmentation_straight.nii -thr 0.5 -o segmentation_straight.nii # in /tmp/sct-20200128160925.047520-label_vertebrae-sny99fmr
Create label to identify disc...
Creating temporary folder...
Run C2-C3 detector...
C2-C3 not detected...
Remove temporary files...
rm -rf /tmp/sct-20200128163454.853102-dty33nlz
Automatic C2-C3 detection failed. Please provide manual label with sct_label_utils
I thus used sct_label_utils with viewer with:
sct_label_utils -i 3dT1.nii -create-viewer 2,3
and it does not give any error. Is that approach right?
thanks,
Rosella
Yes, that’s the right approach
However please make sure the labeling is correct. If it is not, you can bypass this function and just create 2 labels per subjects manually with sct_label_utils as explained in the course.
Sorry but I just today resumed the work on neonatal SC. I have managed to obtain quite a good segmentation results thanks to initial manual set of the centerline. However, I do not know how to create disc label and vertebrae in order to warp PAM50 template to DKI image as sct_label_vertebrae fails to detect C2C3 authomatically . I so used sct_label_utils but my image is very noise and I cannot recognize vertebral discs or levels. How can I handle with this problem?
thanks,
Rosella
Hi Rosella,
You must have acquired a localizer (or any other anatomical image) on which you can clearly identify the discs. You could label the disc on the anat image, and then use it with sct_register_to_template
So I should manually mark the discs using sct_labels-utils in 3dT1 image (the inly structural I have). Register the 3dT1 to the DKI image and then use sct_register_to_template?
Here is an example of my 3dT1 images
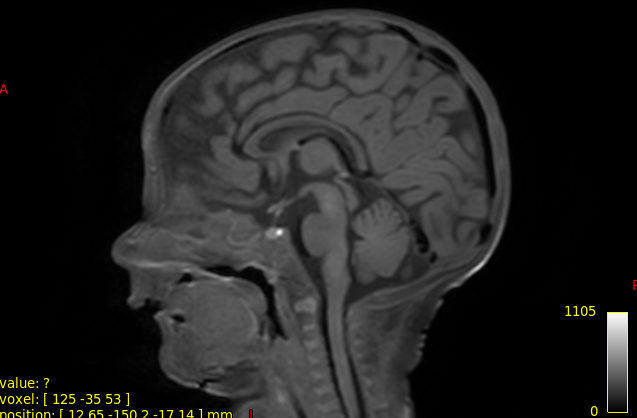
best,
Rosella
you could do the following:
# create two disc labels from the T1w data
sct_label_utils -i T1DATA -create-viewer 3,8 -msg "Create label at the posterior tip of the C2-C3 and C7-T1 discs -o labeldisc_3-8.nii.gz"
# register the PAM50 template to the DWI data. Note, here we use ref subject to prevent straightening (due to the thick slices-- for more info, see SCT course)
sct_register_to_template -i DWI_MEAN -s DWI_MEAN_SEG -ldisc labeldisc_3-8.nii.gz -ref subject
Thank you very much!
Since the FOV in DKI images is reduced between c1 and c3 could I mark disc labels just of that segment?
THanks,
Rosella
Since the FOV in DKI images is reduced between c1 and c3 could I mark disc labels just of that segment?
Yes, absolutely. However, keep in mind that disc c1/c2 is difficult to reliably identify. Maybe try to go for the tip of c1 vertebrae (label 1) and disc c2/c3 (label 3).
sct_label_utils -i T1DATA -create-viewer 1,3 -msg "Create label at the posterior tip of the C1-C2 and C3-C4 discs -o labeldisc_1_3nii.gz"
Should something like this be ok?
best,
Rosella
Moreover, is this ok even if the 3dT1 data and DKI data are not perfeclty registered?
best,
Rosella
sct_label_utils -i T1DATA -create-viewer 1,3 -msg "Create label at the posterior tip of the C1-C2 and C3-C4 discs -o labeldisc_1_3nii.gz
Should something like this be ok?
yes
Moreover, is this ok even if the 3dT1 data and DKI data are not perfeclty registered?
Excellent question. The cord does not need to be perfectly registered in the axial plane, however it needs to match relatively well along the superior-inferior axis. So if there was a repositioning of if the subject moved a lot in the SI direction, you might want to reconsider this approach. You can verify how much the subject moved by overlaying the diffusion scan on the T1 scan
I think there has been movement between T1 and DKI scan. I will attach the 2 images. In case, is it possible to register the DKI image to T1 one (as it has higher resolution)? And warping the template to this registered DKI image?
thanks
Rosella
https://drive.google.com/drive/folders/1kaaz1_zuTsK7dPL4yuR1q4Agm8sH7p17
yes, but i would not recommend it because:
- you are dealing with thick axial slices (hence registration to a higher res will create interpolation errors-- which will bias your qMR metrics extractions)
- you previously mentioned that the discs cannot be seen on the DWI scans, therefore this registration will also be tricky/unreliable
Here is the data.
So should I proceed as we said before, using labels marked on 3dT1?
thnaks
rosella
Hi Rosella,
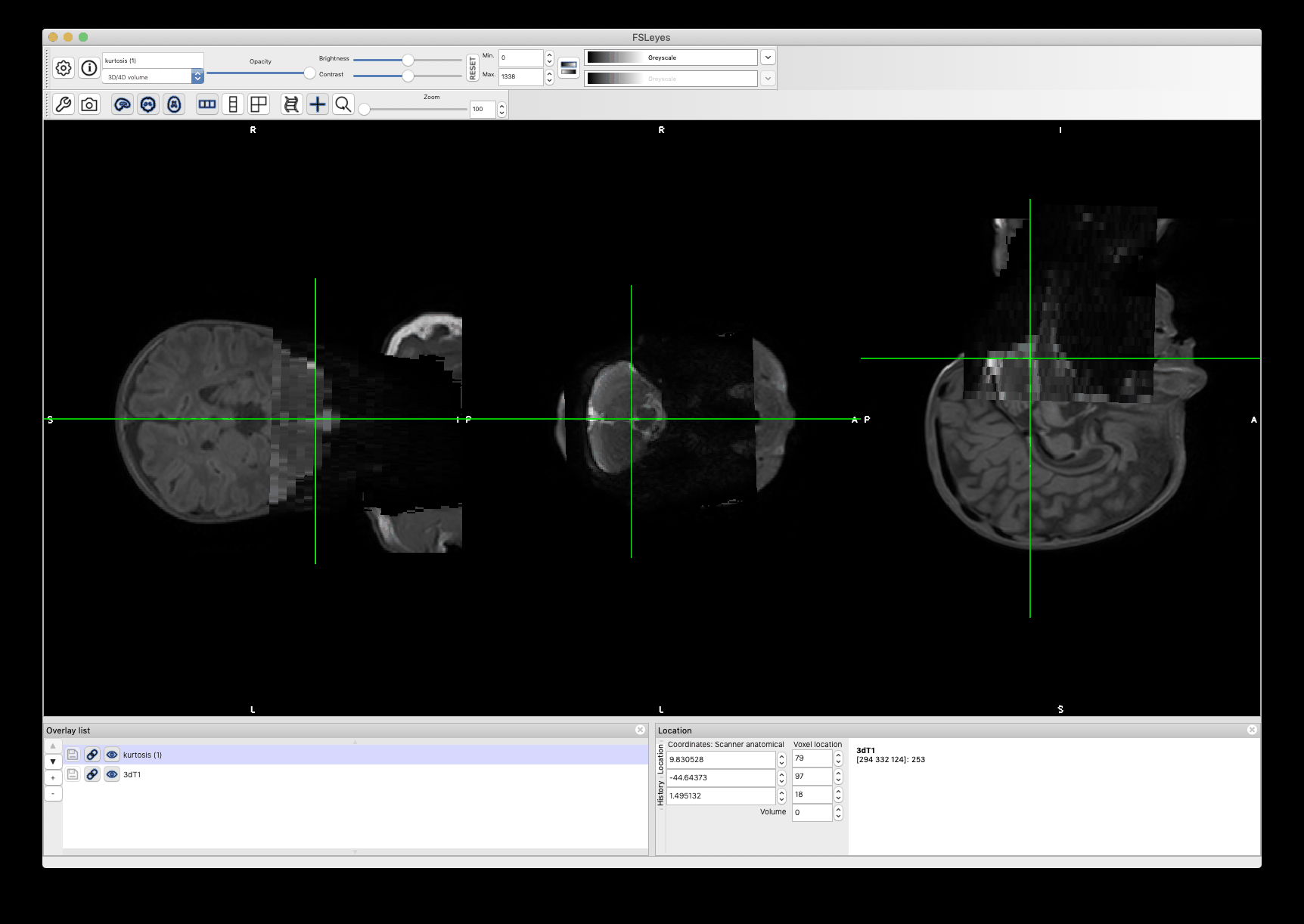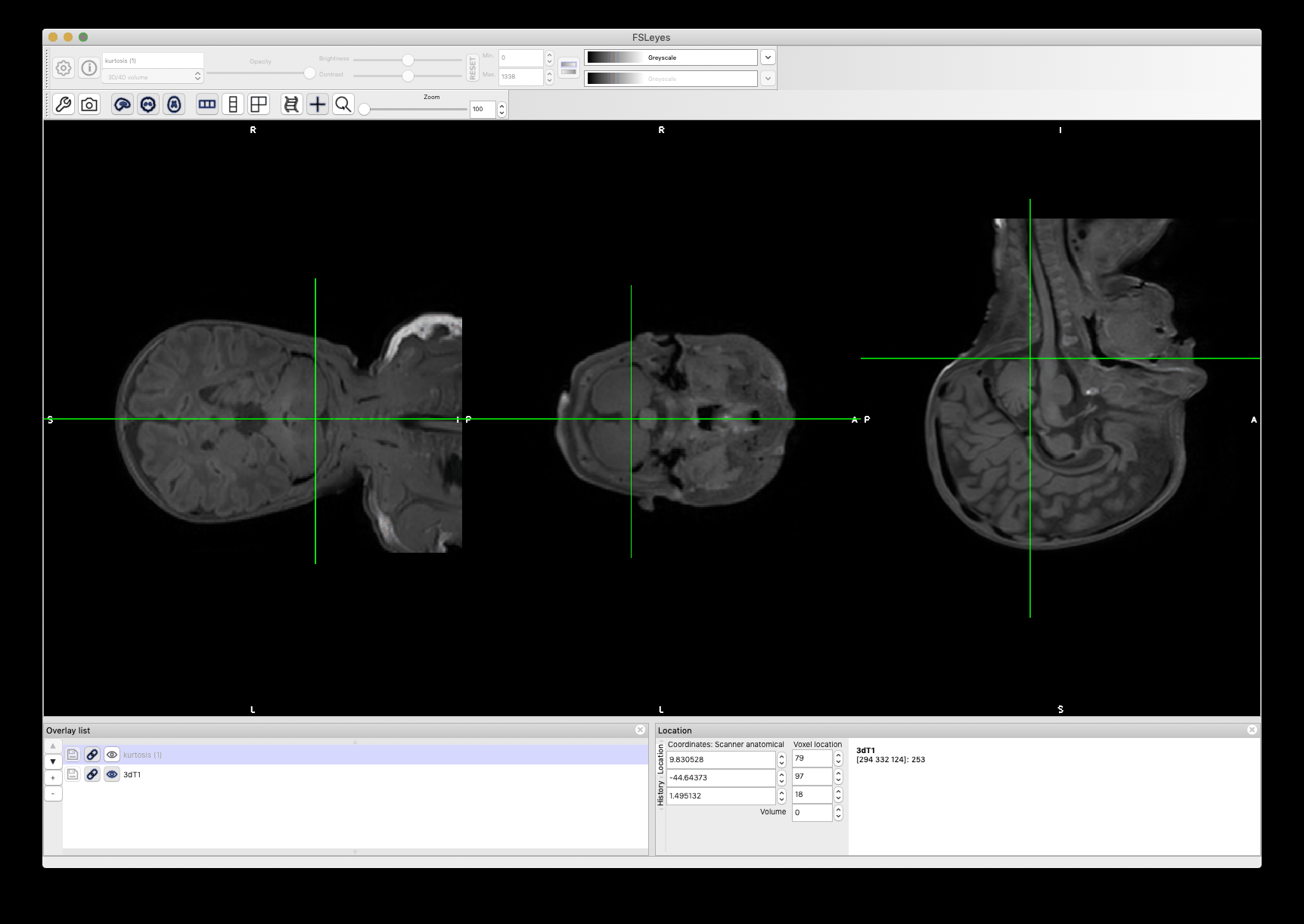
As you can see in the anim below, while both images are clearly not registered along the antero-posterior direction due to the very strong susceptibility artefact, the z-location is similar (see how the bottom tip of the cerebellum is consistent for the two scans). So, since the goal of the disc labeling is only to get a z-value (i.e. along the superior-inferior direction, not in the AP or RL direction), you should probably be OK with labeling from the T1.
Moreover, please keep in mind that this labeling is a very crude approximation of what the spinal level actually is, so even if you would manually label the disc location in the DWI image, that would still be a crude approximation.
Hi. Is there a way to erode a mask of the spinal cord just axially?
best,
Rosella