Hi everyone,
I’m working on diffusion MRI of the cervical and dorsal spinal cord in a cohort of paraplegic and tetraplegic patients, with inflammatory or traumatic spinal cord pathologies. I’m running into an issue with sct_dmri_moco: the command executes without errors, but the output shows no visible motion correction, as if no transformation had been applied.
My dataset consists of HARDI diffusion sequences (Labounek et al., 2020) in two acquisitions: 70 AP and 70 PA volumes. Among these 70 volumes, 63 are diffusion-weighted images (42 directions at b = 1000 s/mm² and 21 directions at b = 550 s/mm²), and the remaining 7 are b0 volumes. The acquisition was performed using interleaved multi-slice mode and an interleaved series, which I noticed was also mentioned as a potential source of issues in a previous thread here on the forum.
For preprocessing, I first applied topup and eddy, for the distortion-correction. I then ran sct_dmri_moco using the topupeddy-corrected data as input, a mask covering about 35 mm around the spinal cord (to include both the cord and surrounding anatomical structures) and bvec of my raw data (vector 3x140). My initial command was:
sct_dmri_moco -i 00246888_dMRI_HARDI_AP_PA_EP_ez_eddy.nii.gz -bvec AP_PA.bvec -m mask_00246888_dMRI_HARDI_AP_PA_EP_ez_eddy_dwi_mean.nii.gz -param metric=MeanSquares
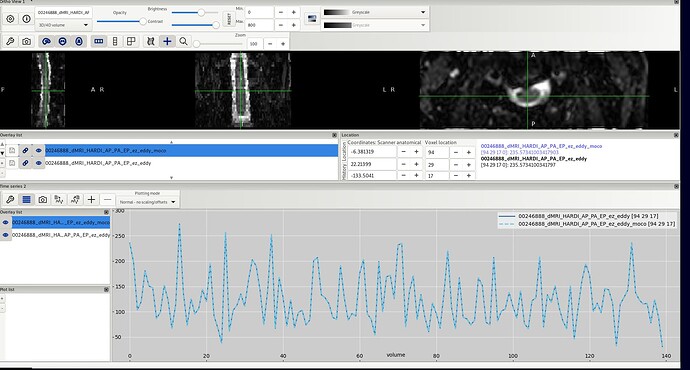
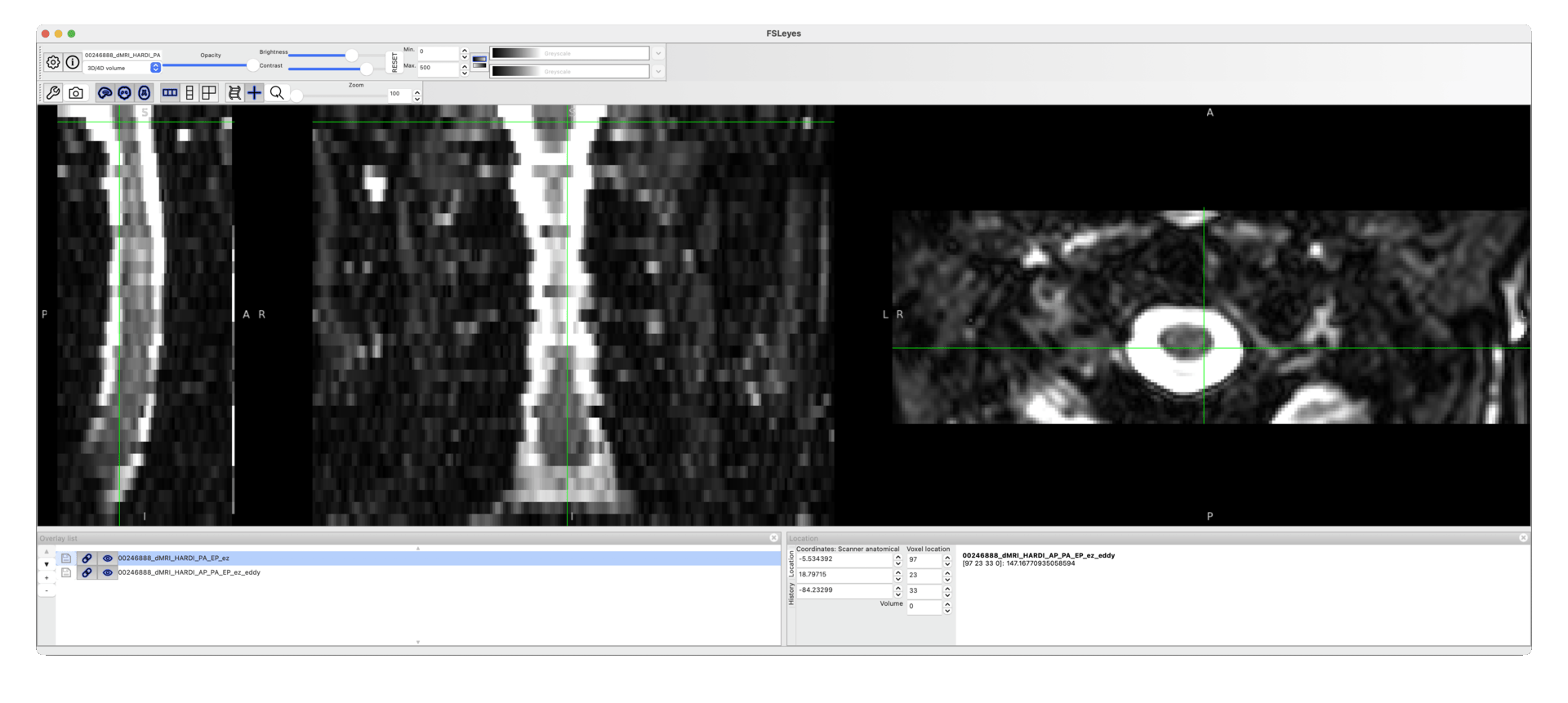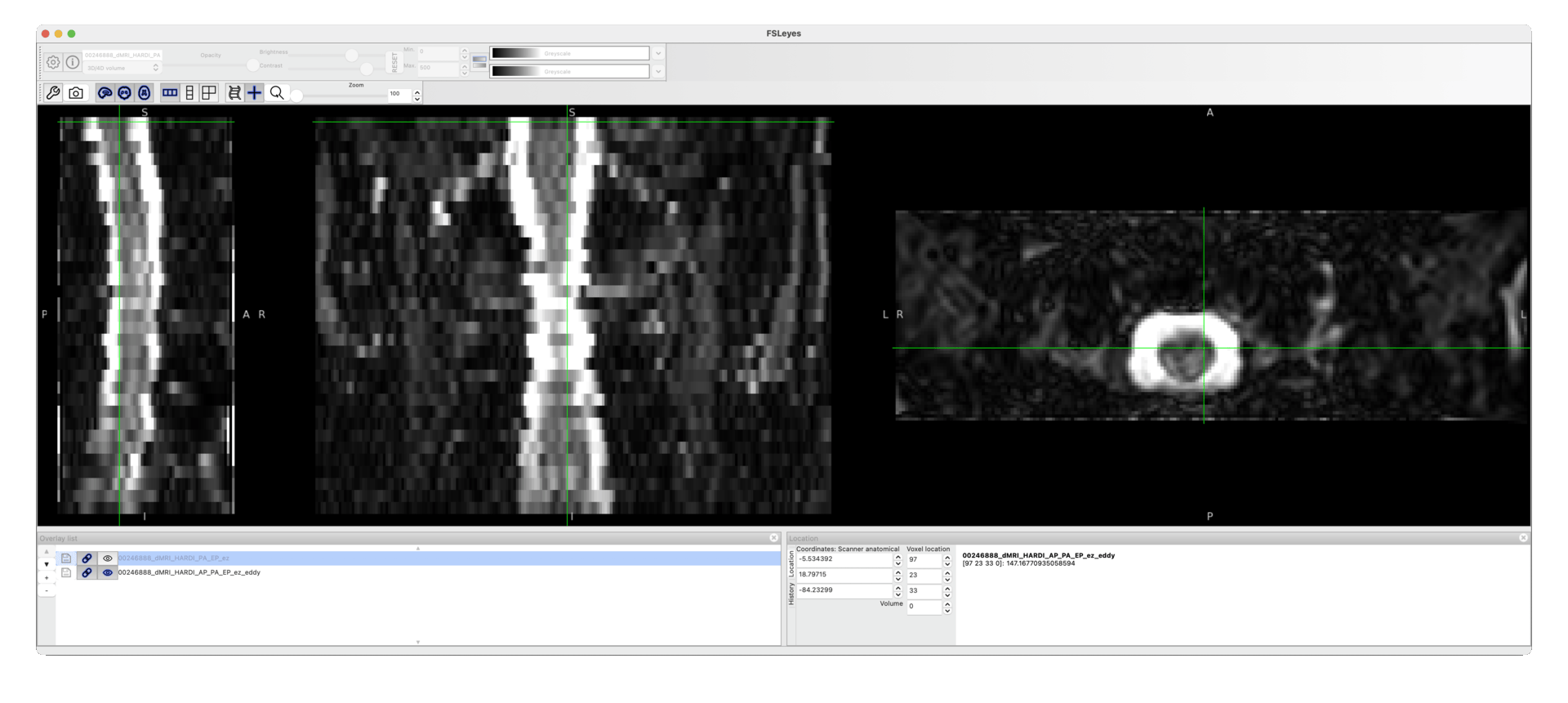
The command runs with no errors, but the resulting data are identical to the input.
I am attaching here a screenshot to show that the output does not differ from the input
To investigate further, I checked this related discussion:
[Sct_dmri_moco doesn't seem to correct motion]
I tried several suggestions from that thread, including setting poly=0, enabling per-volume correction (g=1), increasing the number of iterations, and increasing the gradStep. I also tried running motion correction on AP-only volumes using the rotated bvecs from eddy. Unfortunately, none of these attempts produced any visible correction.
As a control test, I ran sct_dmri_moco directly on the raw data before applying topup and eddy, and in that case motion correction worked.
Do you have any suggestions? Could this be related to the interleaved acquisition or to the interaction between topup/eddy outputs and sct_dmri_moco?
I hope I have been exhaustive in describing the issue.
Thank you very much for your consideration