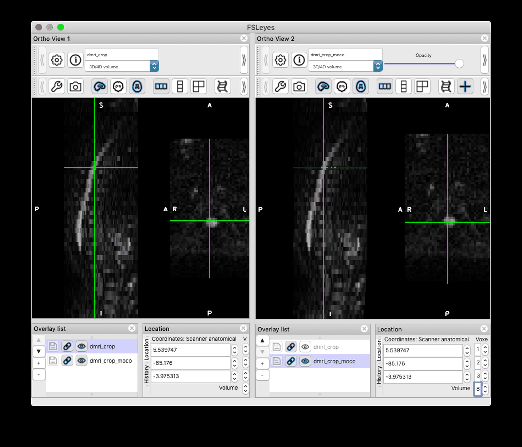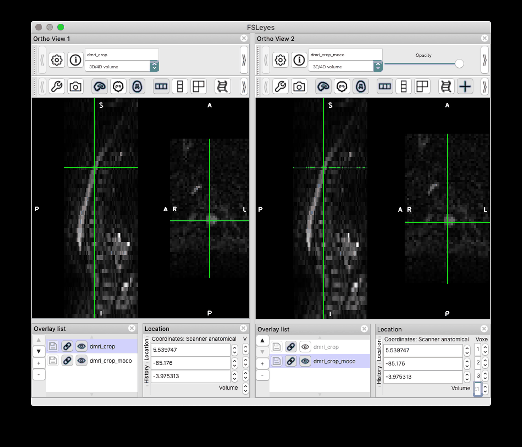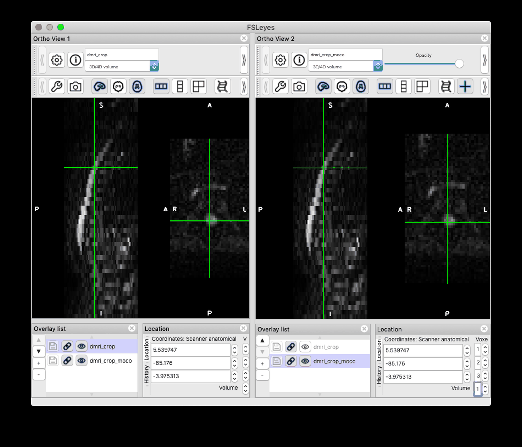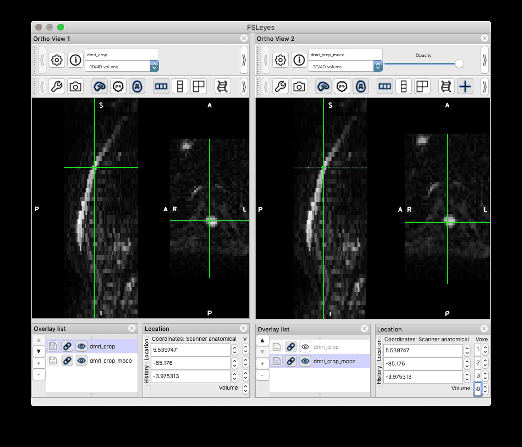
I followed your steps and was able to produce the same results. Thank you so much!! I would like to give some feedback and one related question:
-
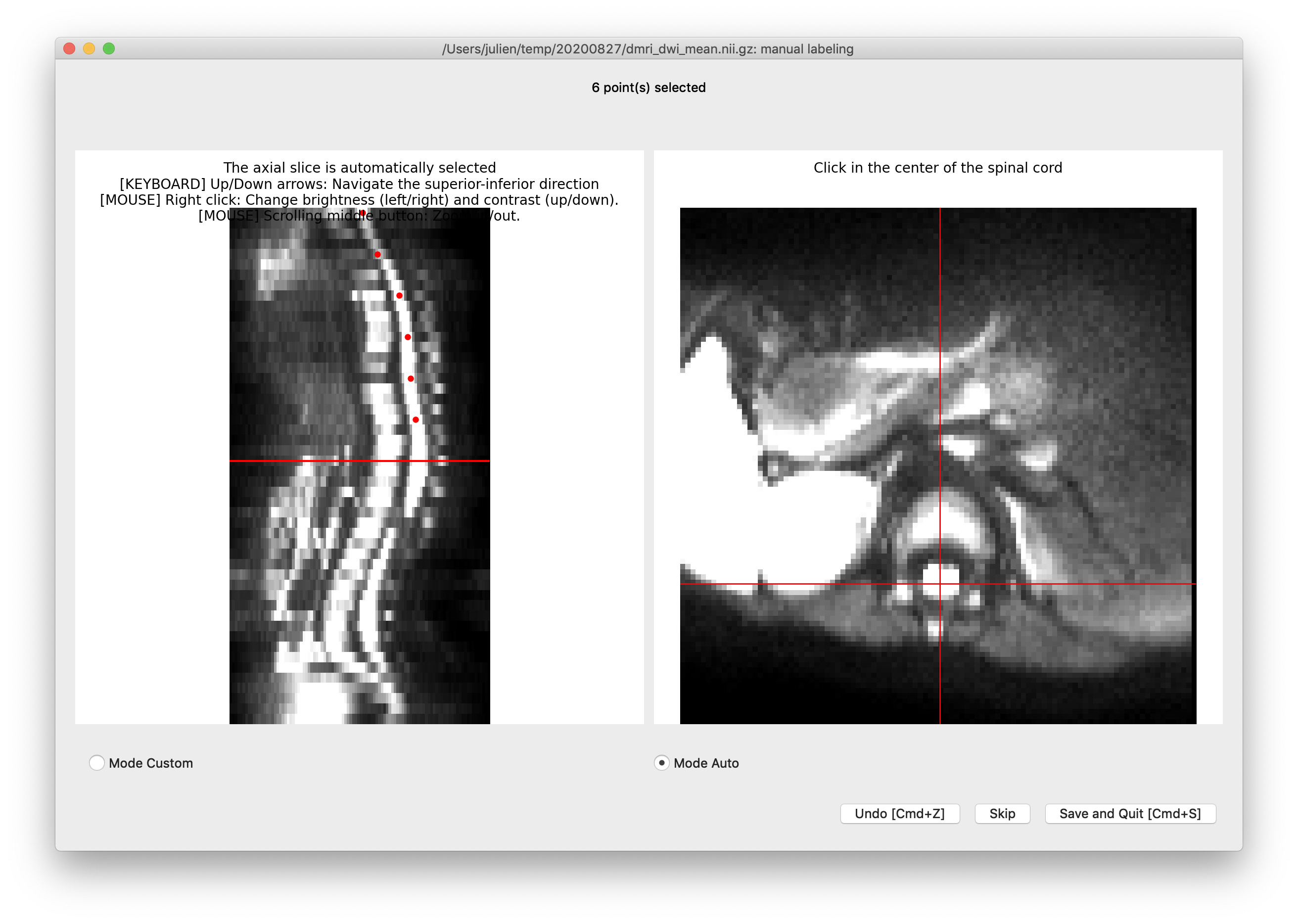
Feedback on the centerline generation. The semi-manual code doesn’t work on my end, with some Qt flatform plugin issue. So I used b0_mean to get the centerline, which seems to be more robust than using dwi_mean. The code I used is:
sct_get_centerline -i dmri_b0_mean.nii.gz -c dwi -method optic -centerline-algo bspline
-
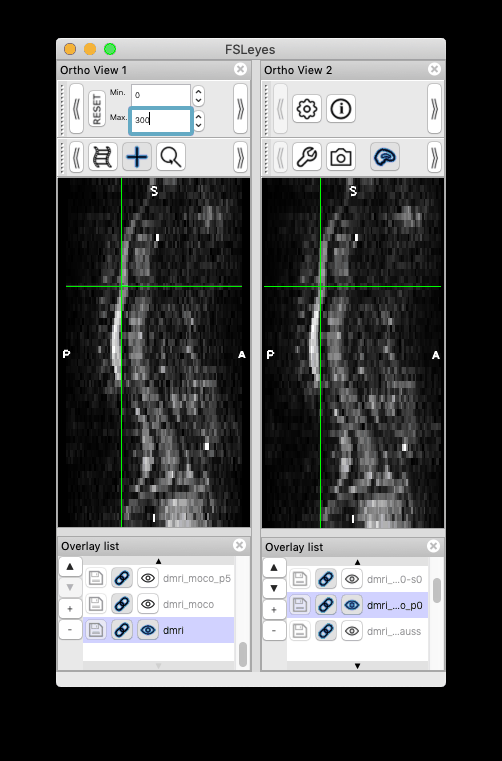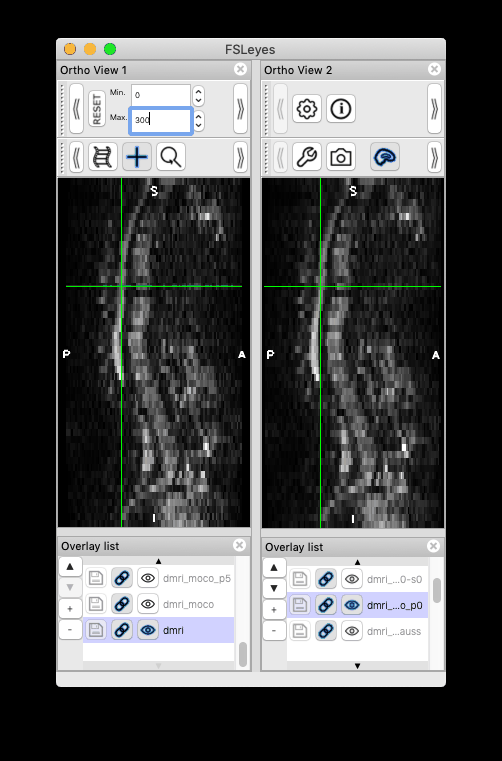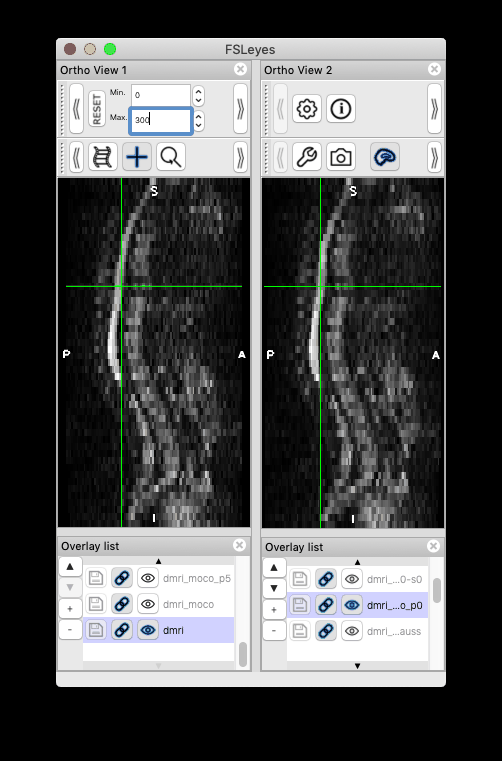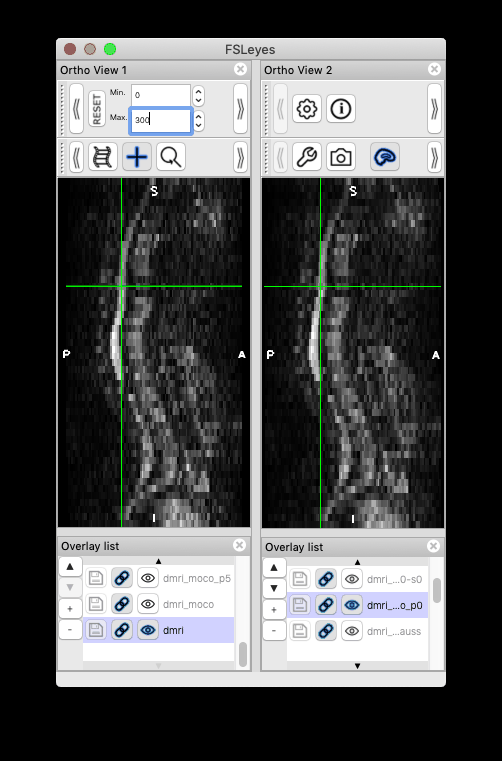
Feeback on version issue: I installed the most recent development version following the instructions in the link you attached (version 4.3). However, this version doesn’t produce the same moco results (i.e. motion persists). Instead, my previous version sct_dev (checked out in March, version 4.2.2) was able to reproduce the same results as you have.
-
Error with other datasets: I processed a few other subjects (with sct_dev 4.2.2). Some of them run through with good results, but some of them had error messages with “No good transformation exist.”. Could you help take a look? Please see below for the data link/ code/output/data link. Thanks ahead of time!
data link:
https://iu.box.com/s/7xntv0nc3ejb60eh64u3f73b2siw458t
Code I used:
Average DWI data
sct_dmri_separate_b0_and_dwi -i dmri.nii.gz -bvec dmri.bvec -a 1
get centerline auto in b0 image
sct_get_centerline -i dmri_b0_mean.nii.gz -c dwi -method optic -centerline-algo bspline
sct_create_mask -i dmri_b0_mean.nii.gz -p centerline,dmri_b0_mean_centerline.nii.gz -size 35mm -f gaussian -o dmri_b0_mean_mask-gauss.nii.gz
Run motion correction.
sct_dmri_moco -i dmri.nii.gz -bvec dmri.bvec -m dmri_b0_mean_mask-gauss.nii.gz -g 1 -param poly=0,metric=MI,smooth=1
Error message:
Spinal Cord Toolbox (4.2.2)
Create temporary folder (/tmp/sct-20200829132202.350591-dmri_moco-_g3td_kv)…
Copying input data to tmp folder and convert to nii…
sct_convert -i /geode2/projects/p060_040/G1/dMRI/DrHo/Spine/20200819_2yo_1.5T_1/processing/dmri/dmri.nii.gz -o /tmp/sct-20200829132202.350591-dmri_moco-_g3td_kv/dmri.nii
cp /geode2/projects/p060_040/G1/dMRI/DrHo/Spine/20200819_2yo_1.5T_1/processing/dmri/dmri.bvec /tmp/sct-20200829132202.350591-dmri_moco-_g3td_kv/bvecs.txt
cp /geode2/projects/p060_040/G1/dMRI/DrHo/Spine/20200819_2yo_1.5T_1/processing/dmri/dmri_b0_mean_mask-gauss.nii.gz /tmp/sct-20200829132202.350591-dmri_moco-_g3td_kv/mask.nii.gz
Get dimensions of data…
100 x 100 x 50
Identify b=0 and DWI images…
WARNING: bvecs file is 3xn instead of nx3. Consider using sct_dmri_transpose_bvecs.
Transpose bvecs…
Number of b=0: 3 [0, 13, 26]
Number of DWI: 36 [1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38]
Split along T dimension…
Merge b=0…
File created: b0.nii
Average b=0…
sct_maths -i b0.nii -o b0_mean.nii -mean t # in /tmp/sct-20200829132202.350591-dmri_moco-_g3td_kv
Merge within groups: 100%|####################| 36/36 [00:50<00:00, 1.40s/iter]
Merging DW files…
Averaging all DW images…
sct_maths -i dwi_averaged_groups.nii -o dwi_averaged_groups.nii_mean.nii.gz -mean t # in /tmp/sct-20200829132202.350591-dmri_moco-_g3td_kv
Estimating motion on b=0 images…
Input parameters:
Input file …b0.nii
Reference file …dmri_T0000.nii.gz
Polynomial degree …1
Smoothing kernel …1
Gradient step …1
Metric …MI
Sampling …0.2
Todo …estimate
Mask …mask.nii.gz
Output mat folder …mat_b0groups
Data dimensions:
100 x 100 x 50 x 3
Copy file_target to a temporary file…
sct_convert -i dmri_T0000.nii.gz -o target.nii.gz
sct_convert -i mask.nii.gz -o mask.nii
Register. Loop across Z (note: there is only one Z if orientation is axial
Z=0/0: 0%| | 0/3 [00:00<?, ?iter/s]WARNING in msct_moco.py: No output. Maybe related to improper calculation of mutual information. Either the mask you provided is too small, or the subject moved a lot. If you see too many messages like this try with a bigger mask. Using previous transformation for this volume (if itexists).
Z=0/0: 33%|############ | 1/3 [00:00<00:00, 3.11iter/s]WARNING in msct_moco.py: No output. Maybe related to improper calculation of mutual information. Either the mask you provided is too small, or the subject moved a lot. If you see too many messages like this try with a bigger mask. Using previous transformation for this volume (if itexists).
Z=0/0: 67%|######################## | 2/3 [00:00<00:00, 3.13iter/s]WARNING in msct_moco.py: No output. Maybe related to improper calculation of mutual information. Either the mask you provided is too small, or the subject moved a lot. If you see too many messages like this try with a bigger mask. Using previous transformation for this volume (if itexists).
Z=0/0: 100%|####################################| 3/3 [00:01<00:00, 2.73iter/s]
ERROR in msct_moco.py: No good transformation exist. Exit program.
(469.0 KB)