Dear SCT Team:
When I was doing gray matter segmentation, the results I got were not ideal.The command I used is as follows:“sct_deepseg_gm -i t2s.nii.gz -qc ~/qc_singleSubj”.
And this is the Sample data:
Sample_Data.zip (9.4 MB)
Hi @jie_Liu,
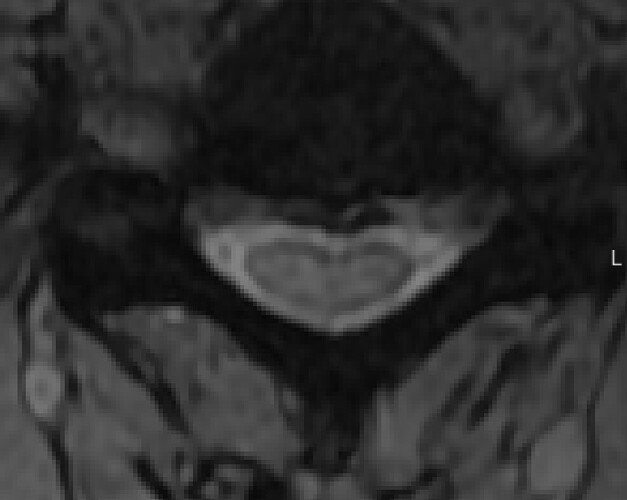
The problem is caused by the image, which does not show the gray matter:
It is not possible for an algorithm to automatically segment something that is not visible in the image. The same goes for segmentation done by a human. I suggest updating the imaging parameters to improve the WM/GM contrast.
Cheers,
Julien
Okay, I understand, thank you very much for your reply.
Hi Julien and SCT team,
I hope it is okay, if I ask a follow-up question to this topic.
I am currently working on segmenting the spinal cord in a dataset of 80 participants, but I am encountering challenges with unsatisfactory segmentation results. Similarly, I suspect these issues may be related to suboptimal contrast between GM, WM, and CSF in the images. Unfortunately, I only have part of the cervical cord, so I hope to get the most out of the data.
Would you have any recommendations on how to enhance the contrast between these structures, either through imaging adjustments or post-processing techniques? I have attached an example of a QC report for your reference.
Kind regard,
Frederik
Hi Frederik,
A couple of things you can do:
- smooth along z (improves CNR, but problematic if strong curvature and thick slices). Example (assuming axial orientation):
sct_maths -i IMAGE -smooth 0,0,3 - smooth along the spinal cord centerline (better, but prone to interpolation error). Example:
sct_smooth_spinalcord -i IMAGE -s SEG - Try the most recent GM segmentation method that @nlaines is currently working on (you could send us 1-2 representative dataset and we can test it on your data)
Hi Julien,
Thank you very much for the reply and suggestions—I truly appreciate it!
It would be a great help if you could review the data. Unfortunately, I’m only permitted to share one example of a healthy control dataset. I’ve been allowed to send the scans of Francesco Grussu, who is helping me with the project. He sends his regards and asks you to take good care of his protons.
Would that work for you?
Have a wonderful weekend!
Kind regards,
Frederik
Sure thing! Say hi back to Francesco ![]()
Hi Julien,
I have shared the data from a onedrive folder but I get a error message, that the invitation is rejected. Do you have any preferred ways that I can share the data safely with you?
Thank you very much for taking the time!
Kind regards,
Frederik
No preferred way. Whatever works for you (eg dropbox, gdrive with password-protected zip)
Great! I have send two datasets in two different email, as I have experienced some issued with the gm segmentation of the first dataset. I hope this is useful for you!
Hi @fnovak,
I didn’t receive any email from you. Here’s my email address: julien.cohen-adad@polymtl.ca
Hi Julien,
I have send another email ![]() I hope it reaches you. Maybe in the spam filter?
I hope it reaches you. Maybe in the spam filter?
my mail is fnovak@cem-cat.org
I got the data and started looking at them. I will come back to you with a solution within a few days.
I appreciate it! thank you
Hi @fnovak , hi @jcohenadad
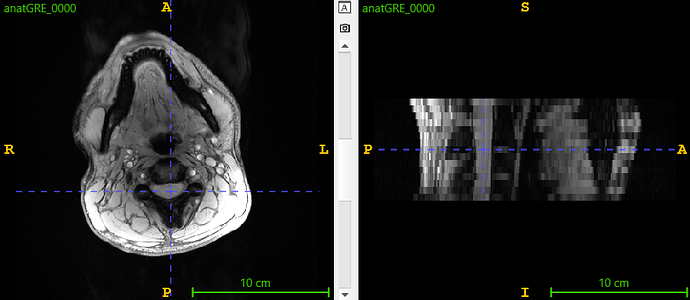
I tried to segment the subject 20340686_696d3c2a-87ff-4f_t2star_degibbs.nii.gz using sct_deepseg_gm, without success (no segmentation output), which surprised me because the GM looks well visible. Here the image:
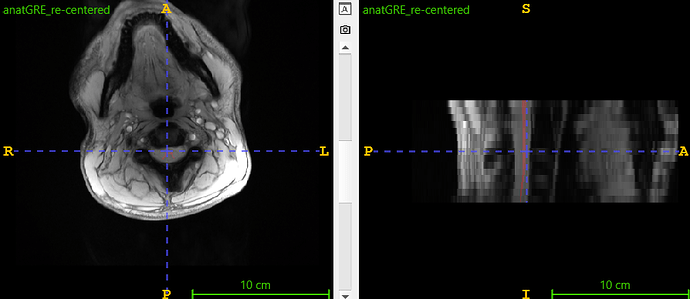
I then found that the SC was off-center. To fix this, I shifted the image 50 pixels along the -Y axis (keeping the same dimensions). This adjustment successfully resulted in segmentation using sct_deepseg_gm. Here the results:
So to ensure that sct_deepseg_gm works, we must make sure that the SC is not too far from the center of the matrix.
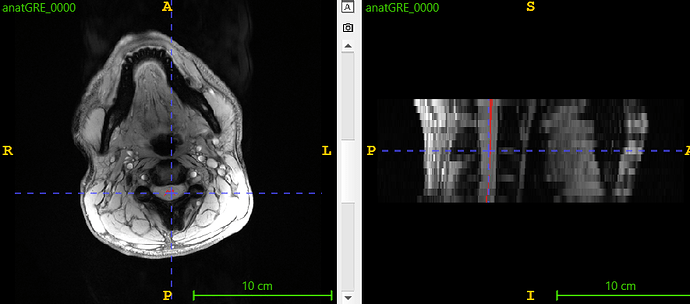
On the other hand, we will soon implement a new model that segments GM even if the images are off-center. Here the results:
Best,
Nilser
To automatize the centering of the cord in the FOV, you could run sct_get_centerline, followed by sct_crop_image. If you need help with this, please let us know.
Hi @fnovak ,
The graymatter model, discussed here, has been implemented in the SCTv.7.0
You can test it on your images with :
sct_deepseg graymatter -i t2s.nii.gz
Best,
Nilser