Hi,
I have been trying to utilize sct_propseg to segment spinal cord and CSF on the following image:
I even tried with -init-centerline viewer (where I basically went through entire spinal cord) and -init-mask viewer, nothing works.
Does anyone know what else I can try in order to get Spinal Cord and corresponding CSF segmentation?
Thanks!
Hi @Stefan_Radonjic if you send me your image I can try to see what’s the best course of action
1 Like
resampled.nii.gz (1.3 MB)
I have resampled image by sct_resample -f 0.5x0.5x0.5 in order to make it small enough to send, so you will probably have to upsample it I guess.
Thanks for help!
downsampling will lower the precision of the image (and upsampling back will not recover the lost information). So you need to send the original data. If size is an issue there are ways like dropbox, gdrive etc
Hi @Stefan_Radonjic ,
So, I confirm that the default sct_propseg commands fails with the default parameters. The key for a robust segmentation is to pass a proper centerline. This could be achieved with sct_deepseg_sc, which segments the cord, and that segmentation could be used as an input centerline to sct_propseg.
Example:
# Segment cord with deepseg
sct_deepseg_sc -i MR_frame_0001.nii.gz -c t2 -centerline svm
# Segment CSF with propseg.
# Note: we specify an output folder in order to avoid overwritting the already existing cord segmentation
sct_propseg -i MR_frame_0001.nii.gz -c t2 -init-centerline MR_frame_0001_seg.nii.gz -CSF -ofolder propseg
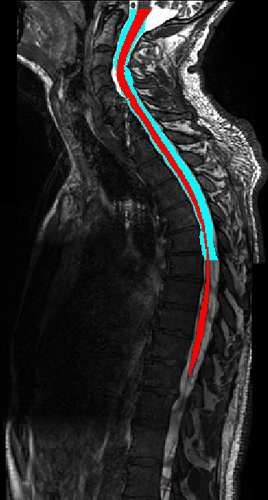
Here is the output segmentation of the cord (red) and CSF (blue):
Note that the CSF segmentation doesn’t go all the way. You might want to play with the propseg parameters to improve this, but at least now you have a good starting point.
2 Likes