Hi @iricchi,
Thank you so much for your question, and for providing great detail (commands run, data). It makes things much easier to troubleshoot, so we very much appreciate it. 
Just to add some additional details summarizing the current issue:
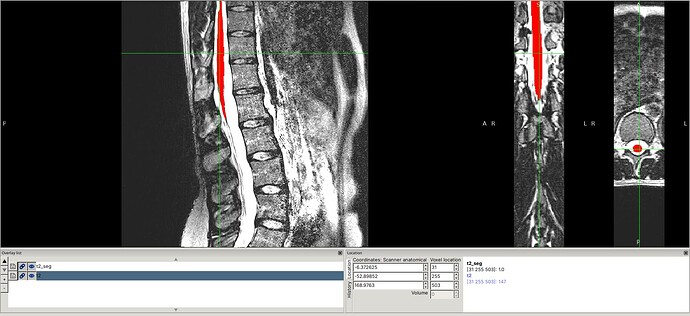
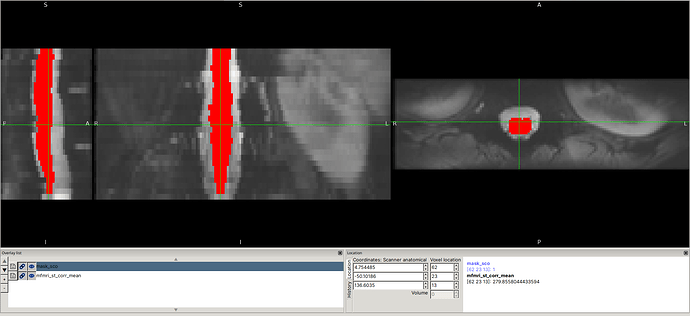
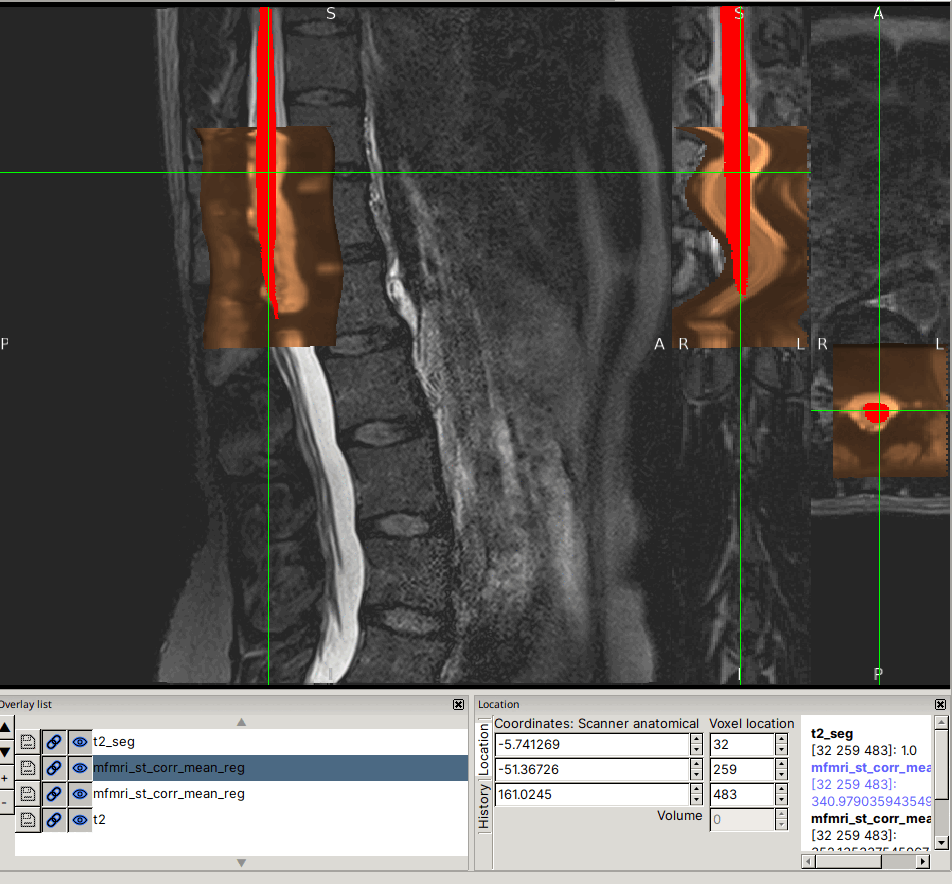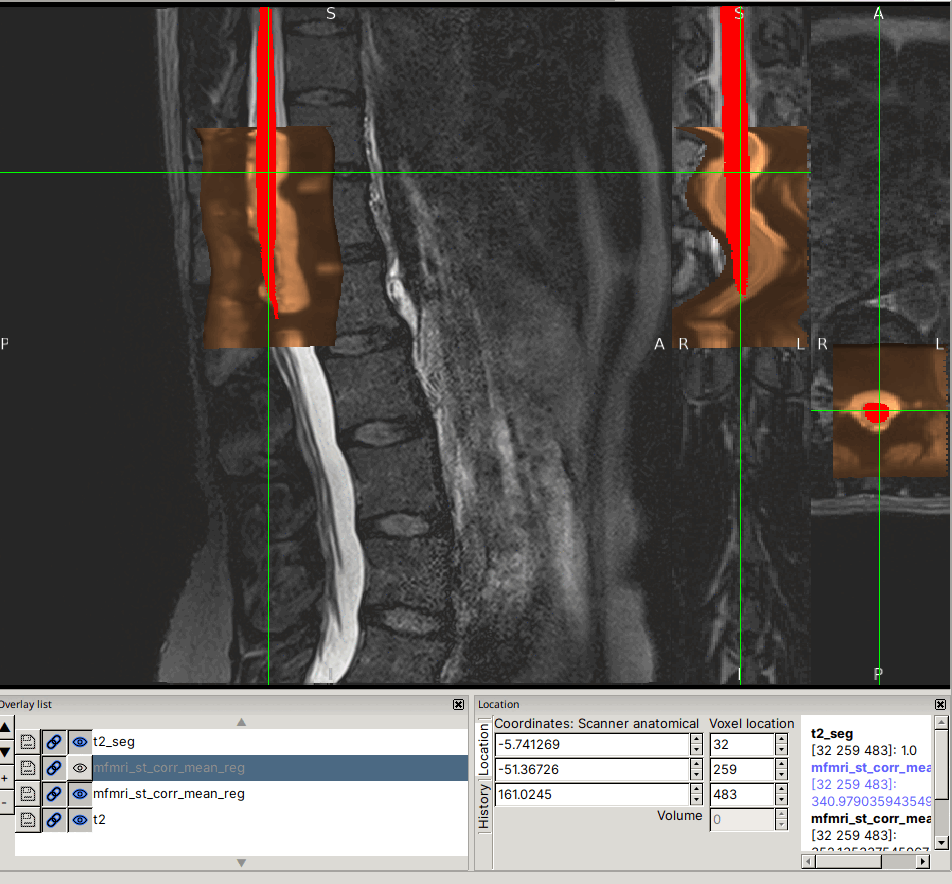
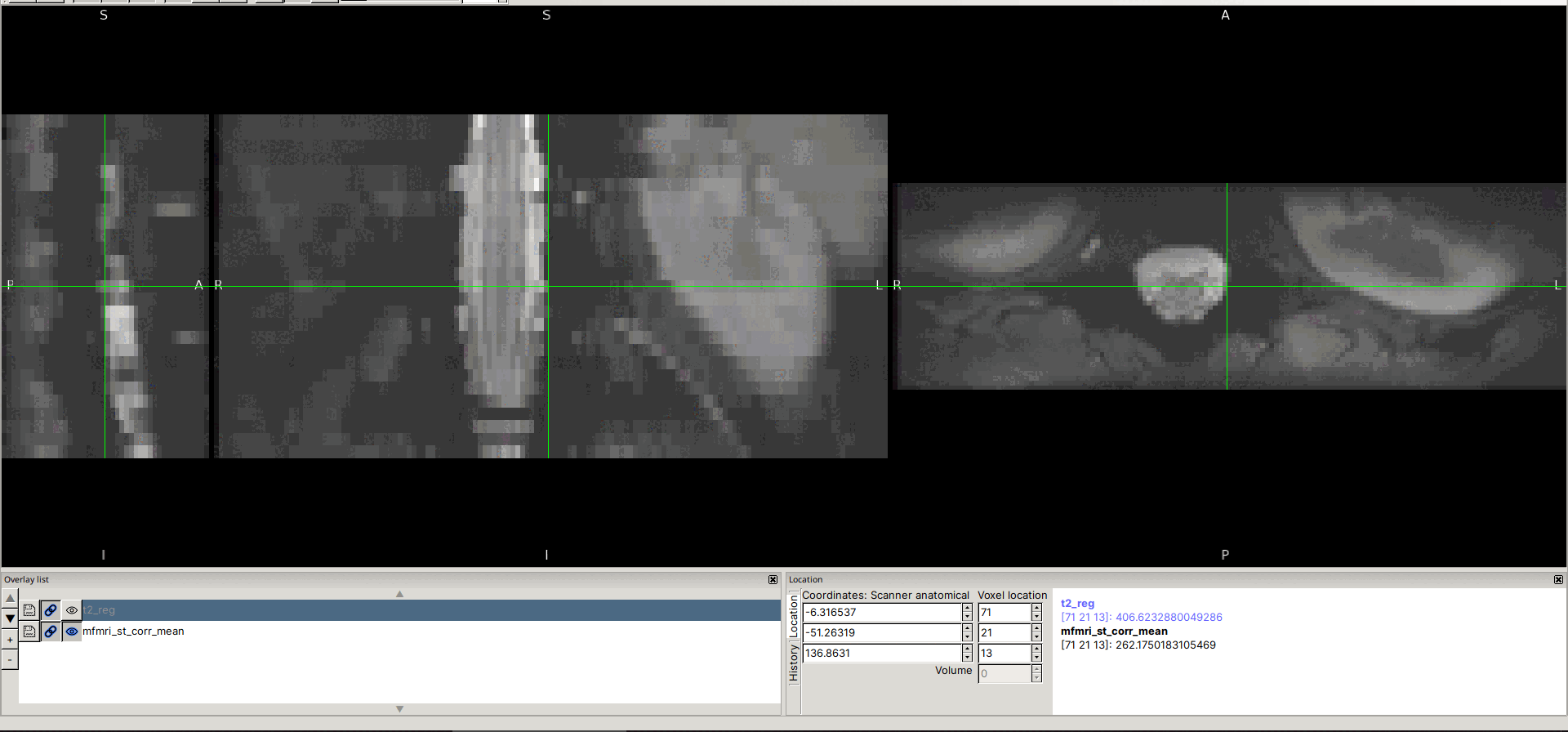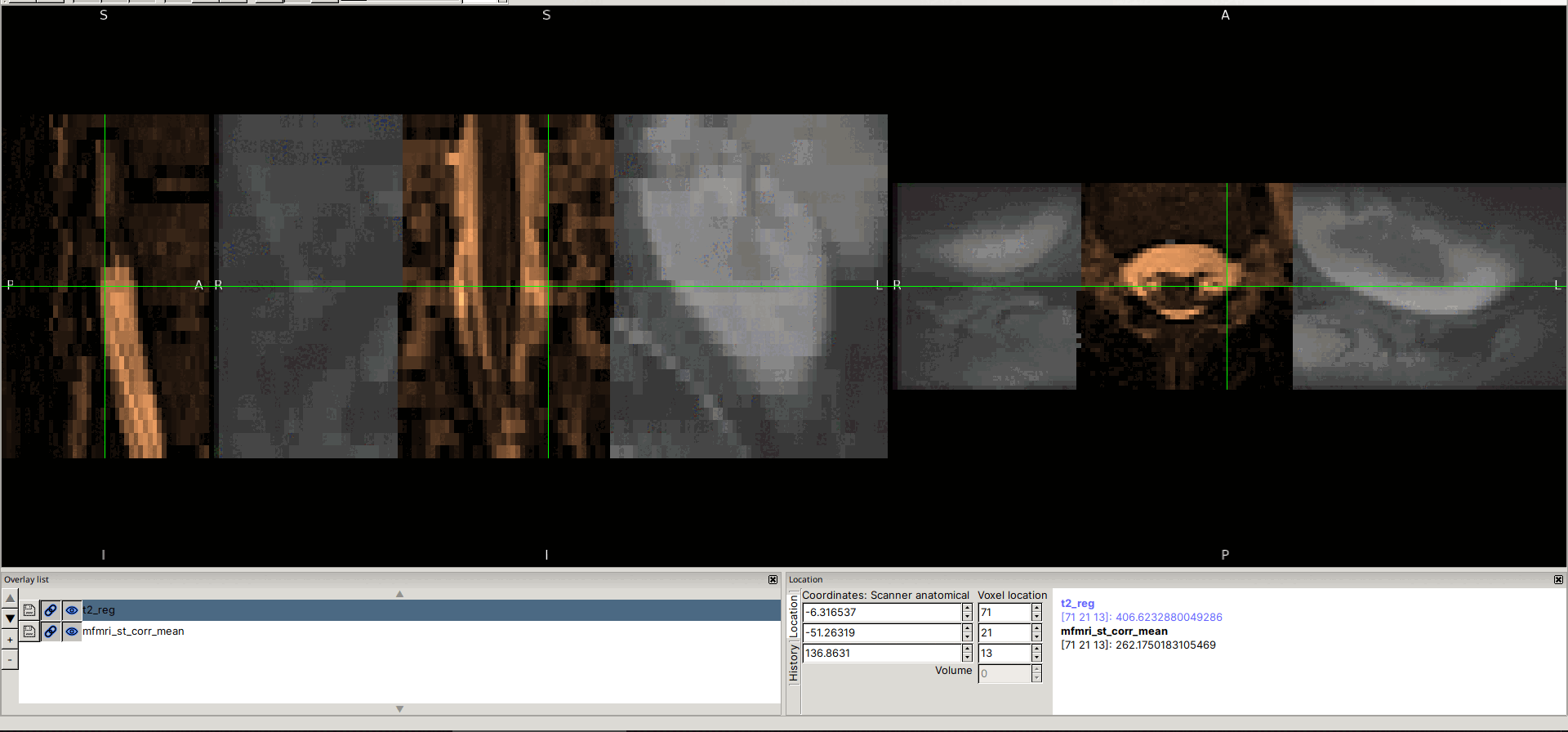
Input data screenshots
t2.nii.gz (with contrast adjusted in viewer):
mfmri_st_corr_mean.nii.gz:
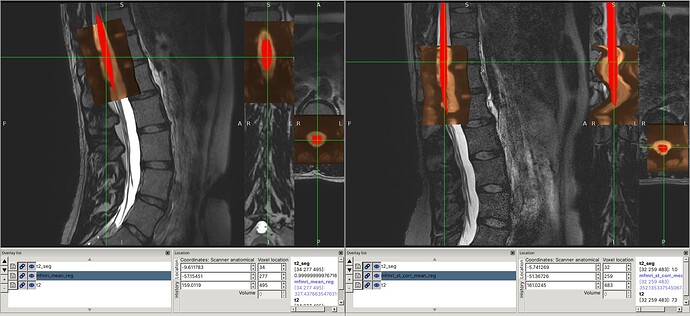
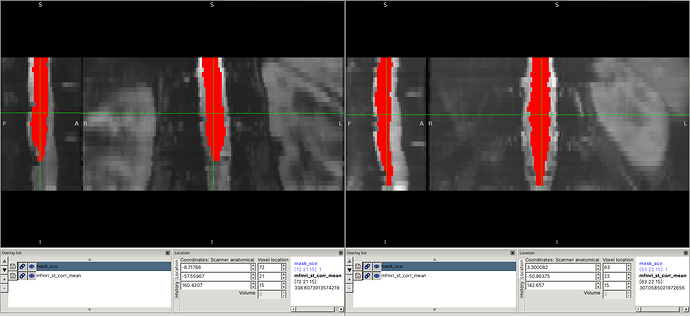
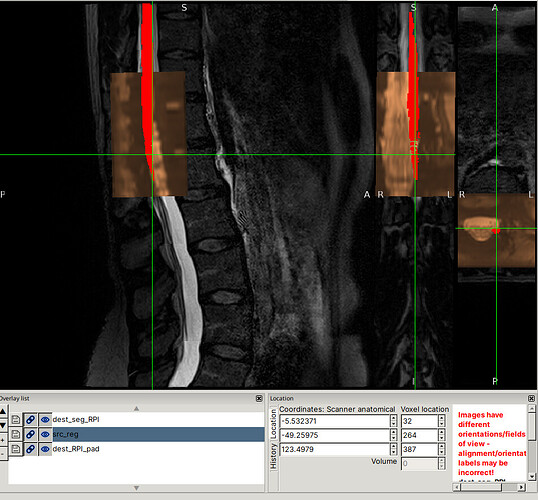
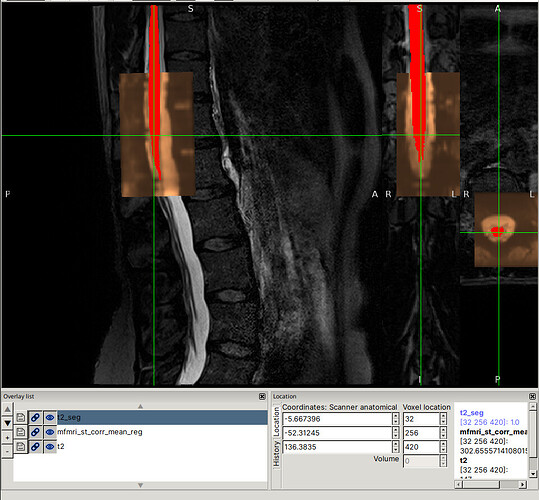
I was able to reproduce the issues you describe using the command you provided (I substituted the file paths so that they would be flat, for use with the files provided in the drive link):
# -param was rearranged for visual clarity
sct_register_multimodal -i mfmri_st_corr_mean.nii.gz \
-d t2.nii.gz \
-iseg mask_sco.nii.gz \
-dseg t2_seg.nii.gz \
-param step=1,type=seg,algo=slicereg,metric=MeanSquares:
step=2,type=seg,algo=affine,metric=MeanSquares,gradStep=0.2:
step=3,type=im,algo=syn,metric=CC,iter=5,shrink=2;
-ofolder test
Full output log
--
Spinal Cord Toolbox (git-master-069fe79472f3e7a664e0e38c4f123b149814aa59)
sct_register_multimodal -i mfmri_st_corr_mean.nii.gz -d t2.nii.gz -iseg mask_sco.nii.gz -dseg t2_seg.nii.gz -param step=1,type=seg,algo=slicereg,metric=MeanSquares:step=2,type=seg,algo=affine,metric=MeanSquares,gradStep=0.2:step=3,type=im,algo=syn,metric=CC,iter=5,shrink=2
--
Input parameters:
Source .............. mfmri_st_corr_mean.nii.gz (144, 44, 27)
Destination ......... t2.nii.gz (64, 640, 640)
Init transfo ........
Mask ................
Output name .........
Remove temp files ... 1
Verbose ............. 1
Check if input data are 3D...
Creating temporary folder (/tmp/sct_2024-05-06_12-05-27_register-wrapper_0m19r2fk)
Copying input data to tmp folder and convert to nii...
--
ESTIMATE TRANSFORMATION FOR STEP #0
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... MI
samplStrategy .. None
samplPercent ... 0.2
iter ........... 0
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/joshua/repos/spinalcordtoolbox/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'MI[dest_RPI.nii,src.nii,1,32]' --convergence 0 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step0,src_regStep0.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2024-05-06_12-05-27_register-wrapper_0m19r2fk
--
ESTIMATE TRANSFORMATION FOR STEP #1
Apply transformation from previous step
/home/joshua/repos/spinalcordtoolbox/bin/isct_antsApplyTransforms -d 3 -i src_seg.nii -o src_seg_reg.nii -t warp_forward_0.nii.gz -r dest_seg_RPI.nii -n NearestNeighbor # in /tmp/sct_2024-05-06_12-05-27_register-wrapper_0m19r2fk
Registration parameters:
type ........... seg
algo ........... slicereg
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
/home/joshua/repos/spinalcordtoolbox/bin/isct_antsSliceRegularizedRegistration -t 'Translation[0.5]' -m 'MeanSquares[dest_seg_RPI_crop.nii,src_seg_reg_crop.nii,1,4,None,0.2]' -p 5 -i 10 -f 1 -s 0 -v 1 -o '[step1,src_seg_reg_crop_regStep1.nii]' # in /tmp/sct_2024-05-06_12-05-27_register-wrapper_0m19r2fk
--
ESTIMATE TRANSFORMATION FOR STEP #2
Apply transformation from previous step
/home/joshua/repos/spinalcordtoolbox/bin/isct_antsApplyTransforms -d 3 -i src_seg.nii -o src_seg_reg.nii -t warp_forward_1.nii.gz warp_forward_0.nii.gz -r dest_seg_RPI.nii -n NearestNeighbor # in /tmp/sct_2024-05-06_12-05-27_register-wrapper_0m19r2fk
Registration parameters:
type ........... seg
algo ........... affine
slicewise ...... 0
metric ......... MeanSquares
samplStrategy .. None
samplPercent ... 0.2
iter ........... 10
smooth ......... 0
laplacian ...... 0
shrink ......... 1
gradStep ....... 0.2
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/joshua/repos/spinalcordtoolbox/bin/isct_antsRegistration --dimensionality 3 --transform 'affine[0.2]' --metric 'MeanSquares[dest_seg_RPI_pad.nii,src_seg_reg.nii,1,4]' --convergence 10 --shrink-factors 1 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step2,src_seg_reg_regStep2.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2024-05-06_12-05-27_register-wrapper_0m19r2fk
--
ESTIMATE TRANSFORMATION FOR STEP #3
Apply transformation from previous step
/home/joshua/repos/spinalcordtoolbox/bin/isct_antsApplyTransforms -d 3 -i src.nii -o src_reg.nii -t warp_forward_2.mat warp_forward_1.nii.gz warp_forward_0.nii.gz -r dest_RPI.nii -n 'BSpline[3]' # in /tmp/sct_2024-05-06_12-05-27_register-wrapper_0m19r2fk
Registration parameters:
type ........... im
algo ........... syn
slicewise ...... 0
metric ......... CC
samplStrategy .. None
samplPercent ... 0.2
iter ........... 5
smooth ......... 0
laplacian ...... 0
shrink ......... 2
gradStep ....... 0.5
deformation .... 1x1x0
init ...........
poly ........... 5
filter_size .... 5
dof ............ Tx_Ty_Tz_Rx_Ry_Rz
smoothWarpXY ... 2
rot_method ..... pca
Estimate transformation
/home/joshua/repos/spinalcordtoolbox/bin/isct_antsRegistration --dimensionality 3 --transform 'syn[0.5,3,0]' --metric 'CC[dest_RPI_pad.nii,src_reg.nii,1,4]' --convergence 5 --shrink-factors 2 --smoothing-sigmas 0mm --restrict-deformation 1x1x0 --output '[step3,src_reg_regStep3.nii]' --interpolation 'BSpline[3]' --verbose 1 # in /tmp/sct_2024-05-06_12-05-27_register-wrapper_0m19r2fk
Concatenate transformations...
/home/joshua/repos/spinalcordtoolbox/bin/isct_ComposeMultiTransform 3 warp_src2dest.nii.gz -R dest.nii warp_forward_3.nii.gz warp_forward_2.mat warp_forward_1.nii.gz warp_forward_0.nii.gz # in /tmp/sct_2024-05-06_12-05-27_register-wrapper_0m19r2fk
/home/joshua/repos/spinalcordtoolbox/bin/isct_ComposeMultiTransform 3 warp_dest2src.nii.gz -R src.nii -i warp_forward_2.mat warp_inverse_0.nii.gz warp_inverse_1.nii.gz warp_inverse_3.nii.gz # in /tmp/sct_2024-05-06_12-05-27_register-wrapper_0m19r2fk
Apply transfo source --> dest...
/home/joshua/repos/spinalcordtoolbox/bin/isct_antsApplyTransforms -d 3 -i src.nii -o src_reg.nii -t warp_src2dest.nii.gz -r dest.nii -n Linear # in /tmp/sct_2024-05-06_12-05-27_register-wrapper_0m19r2fk
Apply transfo dest --> source...
/home/joshua/repos/spinalcordtoolbox/bin/isct_antsApplyTransforms -d 3 -i dest.nii -o dest_reg.nii -t warp_dest2src.nii.gz -r src.nii -n Linear # in /tmp/sct_2024-05-06_12-05-27_register-wrapper_0m19r2fk
Generate output files...
File created: mfmri_st_corr_mean_reg.nii.gz
mv /tmp/sct_2024-05-06_12-05-27_register-wrapper_0m19r2fk/warp_src2dest.nii.gz warp_mfmri_st_corr_mean2t2.nii.gz
File created: warp_mfmri_st_corr_mean2t2.nii.gz
File created: t2_reg.nii.gz
mv /tmp/sct_2024-05-06_12-05-27_register-wrapper_0m19r2fk/warp_dest2src.nii.gz warp_t22mfmri_st_corr_mean.nii.gz
File created: warp_t22mfmri_st_corr_mean.nii.gz
Remove temporary files...
rm -rf /tmp/sct_2024-05-06_12-05-27_register-wrapper_0m19r2fk
Finished! Elapsed time: 1668s
Done! To view results, type:
fsleyes mfmri_st_corr_mean.nii.gz t2_reg.nii.gz &
Done! To view results, type:
fsleyes t2.nii.gz mfmri_st_corr_mean_reg.nii.gz &
Note for @jcohenadad: This command took quite a long time on my laptop (almost 30 minutes), with most of the time spent on Step 3, likely due to metric=CC (which is known to take quite a bit of time). I’m mentioning this ahead of time so that we can appropriately plan for testing other commands.
I am not sure why this is the case specifically for this subject.
Given that your command has been working well for you for other subjects, could you possibly share the data for one of the subjects where there are no issues? (It might help us to compare the input data to see if there are any important differences with this subject in particular.)
Kind regards,
Joshua