Dear SCT team,
We are having some problems with the registration of epi data to the PAM50 template.
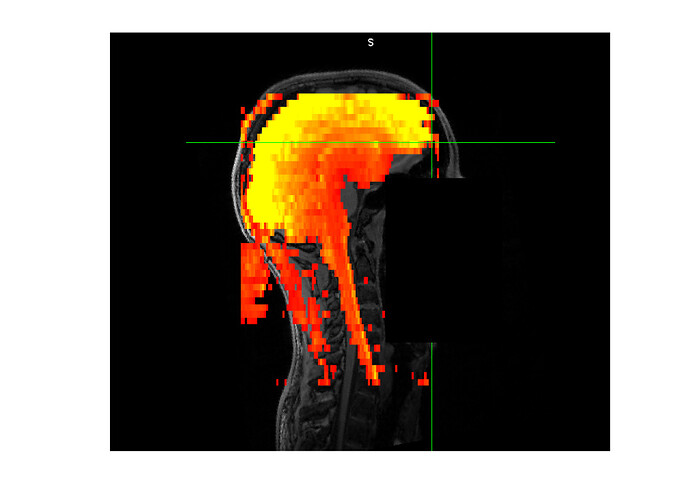
Please find attached an example of our fMRI data (red-yellow) on top of the subject’s T1 (greyscale). The brain is in reasonable good alignment but you can see how the end of the spinal cord in the epi massively deviates from the T1.
We register the T1 to the PAM50 template using the standard SCT pipeline suggested in the manual and it works perfectly.
Here is the command that we use to register the epi to the template:
sct_register_multimodal -i $SCT_DIR/data/PAM50/template/PAM50_t1.nii.gz -d epi_moco_mean.nii.gz -iseg $SCT_DIR/data/PAM50/template/PAM50_cord.nii.gz -dseg epi_moco_mean_seg.nii.gz -param step=1,type=seg,algo=slicereg,smooth=3:step=2,type=seg,algo=centermass,slicewise=1,iter=3 -initwarp warp_template2anat.nii.gz -initwarpinv warp_anat2template.nii.gz
this is what we found to be the best set of parameters for our data, but we are not very happy with the results and we were wondering if you believe that given the distortion of our images this is a good approach and/or if you had any other suggestion (e.g. increasing the permitted deformation)?
Thank you,
Valeria
Hi Valeria,
the first thing to check would be the EPI segmentation (epi_moco_mean_seg.nii.gz). If you added the -qc flag in your analysis, can you zip the qc report and send it to me so I can see where things go wrong?
Best
Julien
Hi Julien,
Thank you for your prompt response and apologies for my delay.
I segmented the cord with deepseg and then adjusted the segmentation manually so it should be ok.
I attached the quality check for the epi registration to template. You will see that the last slices are misaligned to the template. This is a problem but actually our main concern is the misalignment of the discs in the z-direction.
All the best,
Valeria
qc_epi_reg.zip (362.4 KB)
Hi Valeria,
I would use the T2 (or T2*) contrast of the template for registration, because your EPI is T2*-weighted. Also, maybe you could try to be a bit more aggressive with the image-based registration. Try those different combinations of parameters, then send me the qc so we can compare:
sct_register_multimodal -i $SCT_DIR/data/PAM50/template/PAM50_t2s.nii.gz -d epi_moco_mean.nii.gz -iseg $SCT_DIR/data/PAM50/template/PAM50_cord.nii.gz -dseg epi_moco_mean_seg.nii.gz -param step=1,type=seg,algo=slicereg,smooth=3:step=2,type=seg,algo=centermass,slicewise=1,iter=3 -initwarp warp_template2anat.nii.gz -initwarpinv warp_anat2template.nii.gz -qc qc_reg2template
sct_register_multimodal -i $SCT_DIR/data/PAM50/template/PAM50_t2s.nii.gz -d epi_moco_mean.nii.gz -iseg $SCT_DIR/data/PAM50/template/PAM50_cord.nii.gz -dseg epi_moco_mean_seg.nii.gz -param step=1,type=seg,algo=centermass:step=2,type=seg,algo=bsplinesyn,metric=MeanSquares,slicewise=1,smooth=0,iter=3 -initwarp warp_template2anat.nii.gz -initwarpinv warp_anat2template.nii.gz -qc qc_reg2template
sct_register_multimodal -i $SCT_DIR/data/PAM50/template/PAM50_t2s.nii.gz -d epi_moco_mean.nii.gz -iseg $SCT_DIR/data/PAM50/template/PAM50_cord.nii.gz -dseg epi_moco_mean_seg.nii.gz -param step=1,type=seg,algo=centermass:step=2,type=im,algo=syn,metric=CC,slicewise=1,smooth=0,iter=3 -initwarp warp_template2anat.nii.gz -initwarpinv warp_anat2template.nii.gz -qc qc_reg2template
As for the mis-alignment in the z-direction, there is not much we can do, apart from redefining the disc levels on the EPI space. You don’t want to apply a z-translation of your EPI data because it would cause too much interpolation errors (thick slices).
Cheers,
Julien
Hi Julien,
I attached the new qc with your combination of parameters. The first two seem actually pretty good, thanks a lot for your help.
I tried defining the discs on the EPI (with sct_label_utils -create-viewer) but after registration they still seem not in perfect alignment with the template, and this will be problematic for group analyses. Is there anything else you would suggest?
All the best,
Valeria
qc_reg2template.zip (452.6 KB)
Hi Valeria,
The 2nd seems reasonably good indeed.
As for the z-alignment, when you say “they still seem not in perfect alignment with the template”, how do you assess the alignment? If you do a label-based method, the z-scaling (as part of register multimodal) should perfectly match the source and destination labels.
In general, if the subject did not move in the superior-inferior direction in the scanner, there is no reason for adjusting the z-scaling with respect to the anatomical image. The distortions you are observing are along the phase-encoding direction (A-P).
Hi Julien,
I’m using the following command to register to template using the disc labels created manually:
sct_register_multimodal -i $SCT_DIR/data/PAM50/template/PAM50_t2s.nii.gz -d epi_moco_mean.nii.gz -iseg $SCT_DIR/data/PAM50/template/PAM50_cord.nii.gz -dseg epi_moco_mean_seg.nii.gz -param step=1,type=seg,algo=centermass:step=2,type=seg,algo=bsplinesyn,metric=MeanSquares,slicewise=1,smooth=0,iter=3 -initwarp warp_template2anat.nii.gz -initwarpinv warp_anat2template.nii.gz -ilabel $SCT_DIR/data/PAM50/template/PAM50_label_discPosterior.nii.gz -dlabel labels.nii.gz
Then I’m overlaying the registered image on top of the T1 PAM50 (assuming that it’s in the same space as the T2s). As you can see in the attached image the discs are not in good alignment.
Best,
Valeria
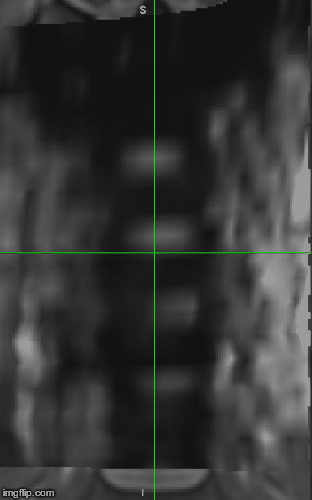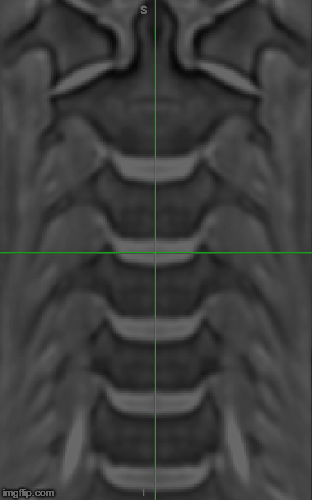
Hi Valeria,
I would recommend checking alignment on the sagittal (not coronal) view, because the registration is done by aligning the posterior tips of the discs and it is difficult to figure out what AP location is shown in the coronal view.
Also please note that the ilabel flag is not made for aligning discs: it is solely for the purpose of initial registration (which is canceled out by the Usenet of initwarp). If you already have manual discs, the use sct_register_to_template without going through the T1.
If it’s easier, I’m happy to write up a personalized batch script if you send me a representative dataset.
Hi Julien,
Unfortunately in the sagittal view the discs look just as bad and sct_register_to_template looks even worse.
It would be great if you could have a go!
Here is a link to an example T1 and epi: https://uob-my.sharepoint.com/:u:/g/personal/vo16427_bristol_ac_uk/Ef0KbNcKTPdBmyIDIIe0iPsBQx9Lg-IuZLN8aW4iaB7Khg?e=zZMwFY
Best,
Valeria
Hi Valeria,
I confirm that due to the large distortions on the EPI, the registration to template via the anatomical image does not yield accurate disc registration. I propose the alternative approach below, which does not rely on the anatomical image, but requires to manually label the levels of two intervertebral discs:
# Segment spinal cord on mean EPI
sct_deepseg_sc -i epi_moco_mean.nii.gz -c t2s -qc qc
# Label at C2/C3 and C6/C7 discs (here we specify the known Z, which can vary across subjects)
sct_label_utils -i epi_moco_mean_seg.nii.gz -create-seg 12,3:0,7 -o labels_disc.nii.gz
# Register to template
sct_register_to_template -i epi_moco_mean.nii.gz -s epi_moco_mean_seg.nii.gz -ldisc labels_disc.nii.gz -c t2s -ref subject -param step=1,type=seg,algo=centermass:step=2,type=im,algo=syn,metric=CC,slicewise=1,smooth=0,iter=3 -qc qc
# Rename warping fields
mv warp_template2anat.nii.gz warp_template2epi.nii.gz
mv warp_anat2template.nii.gz warp_epi2template.nii.gz
# Warp template to EPI space
sct_warp_template -d epi_moco_mean.nii.gz -w warp_template2epi.nii.gz -ofolder label_epi -a 0
Here is the result:
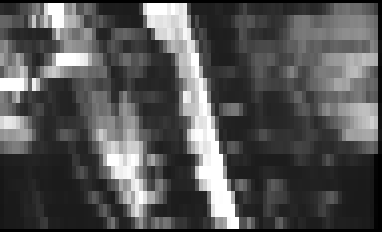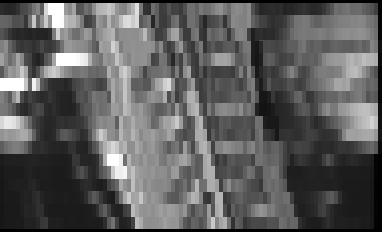
And the QC report: qc.zip (395.6 KB)
If you wish to manually-label discs using sct_label_utils, here is the command (here for C2/C3 and C6/C7 discs):
sct_label_utils -i epi_moco_mean.nii.gz -create-viewer 3,7 -o labels_disc.nii.gz
Hi Julien,
Thanks a lot for this, it seems to work much better. I’ll try the same pipeline on the other subjects and will get in touch again in case I had some other problems.
All the best,
Valeria