Hi,
We are trying to register PAM50 template to our axial T2star data using sct_register_to_template. Since we need properly registered template/PAM50_levels.nii.gz, we use -lspinal flag and levels created by sct_label_utils.
Moreover, we use WM segmentation to take into account both GM and SC shape (flag -s-template-id 4).
Part of our pipeline is:
# Create WM segmentation from SC and GM masks
sct_maths -i sub-0001_ses-01_T2star_seg.nii.gz -sub sub-0001_ses-01_T2star_gmseg.nii.gz -o sub-0001_ses-01_T2star_wmseg.nii.gz
sct_maths -i sub-0001_ses-01_T2star_wmseg.nii.gz -o sub-0001_ses-01_T2star_wmseg.nii.gz -thr 0
# Create mid-vertebral levels. These labels are needed for template registration.
sct_label_utils -i sub-0001_ses-01_T2star_seg_labeled.nii.gz -vert-body 3,4,5,6,7 -o sub-0001_ses-01_T2star_labels_vert.nii.gz
# Register PAM50 template to t2star file using labels created in previous step
sct_register_to_template -i sub-0001_ses-01_T2star.nii.gz -s sub-0001_ses-01_T2star_seg_wmseg.nii.gz -lspinal sub-0001_ses-01_T2star_labels_vert.nii.gz -s-template-id 4 -ref subject -c t2s -param step=1,type=seg,algo=centermassrot,smooth=0:step=2,type=seg,algo=columnwise,smooth=0,smoothWarpXY=2
# Warp template without the white matter atlas
sct_warp_template -d sub-0001_ses-01_T2star.nii.gz -w warp_template2anat.nii.gz -a 0 -ofolder label_t2star
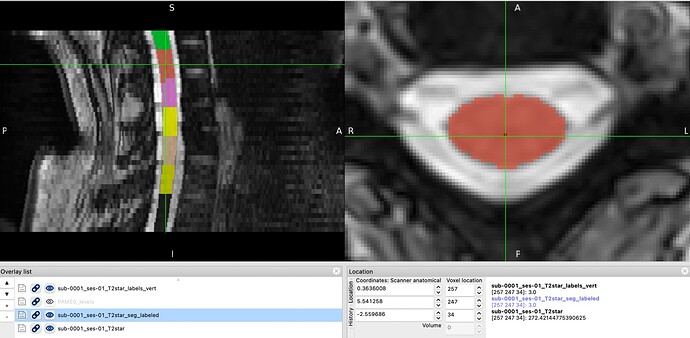
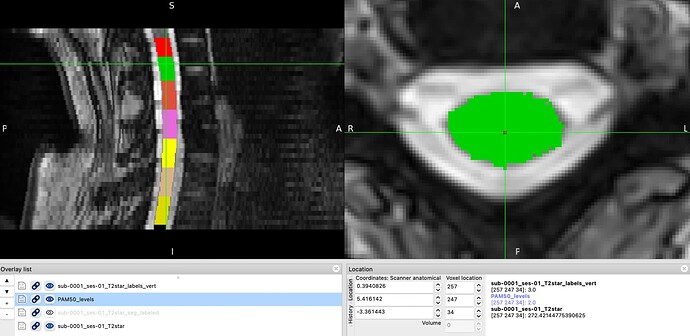
The resulting label_t2star/template/PAM50_levels.nii.gz however does not correspond with original vertebral levels, see images below.
T2star_seg_labeled and T2star_labels_vert (the cursor is placed to the red point with the value 3):
template/PAM50_levels and T2star_labels_vert (the cursor is still placed to the same red point with the value 3, but PAM50_levels has value 2).
In addition, PAM50_wm does not correspond well with the actual WM segmentation.
T2star:
T2star_gmseg:
T2star_wmseg:
template/PAM50_wm and T2star_gmseg:
Here we assumed that PAM50_wm would be alligned to T2star_wmseg.
SCT info:
sct_check_dependencies -short
--
SCT info:
- version: git-master-afdb05da537a75a34f3434f5512473f4e34f55bf*
Link to our data:
Thank you!
Jan and René