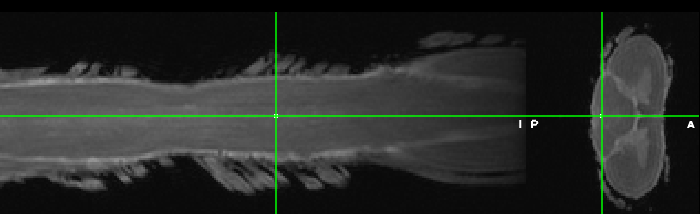
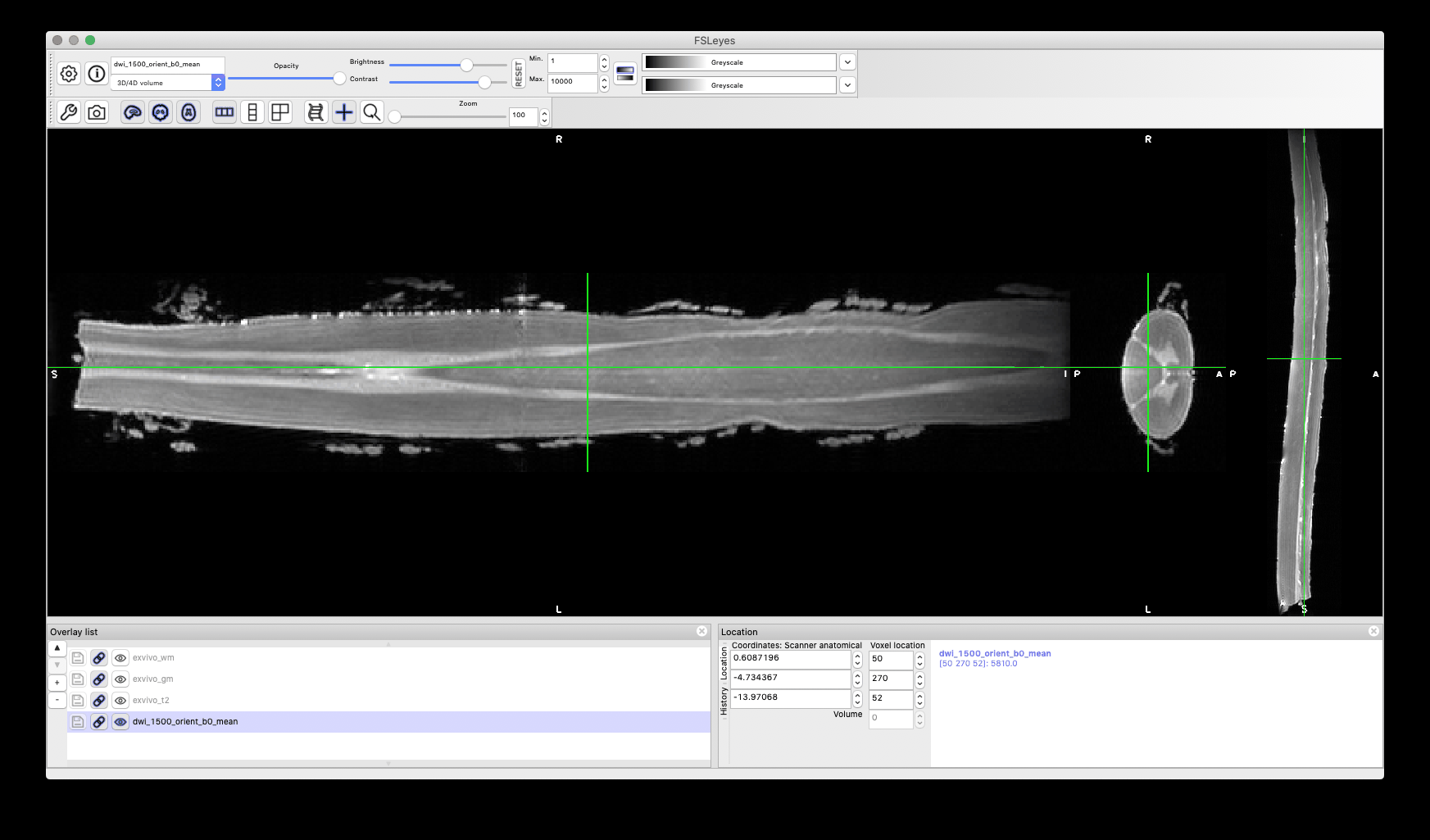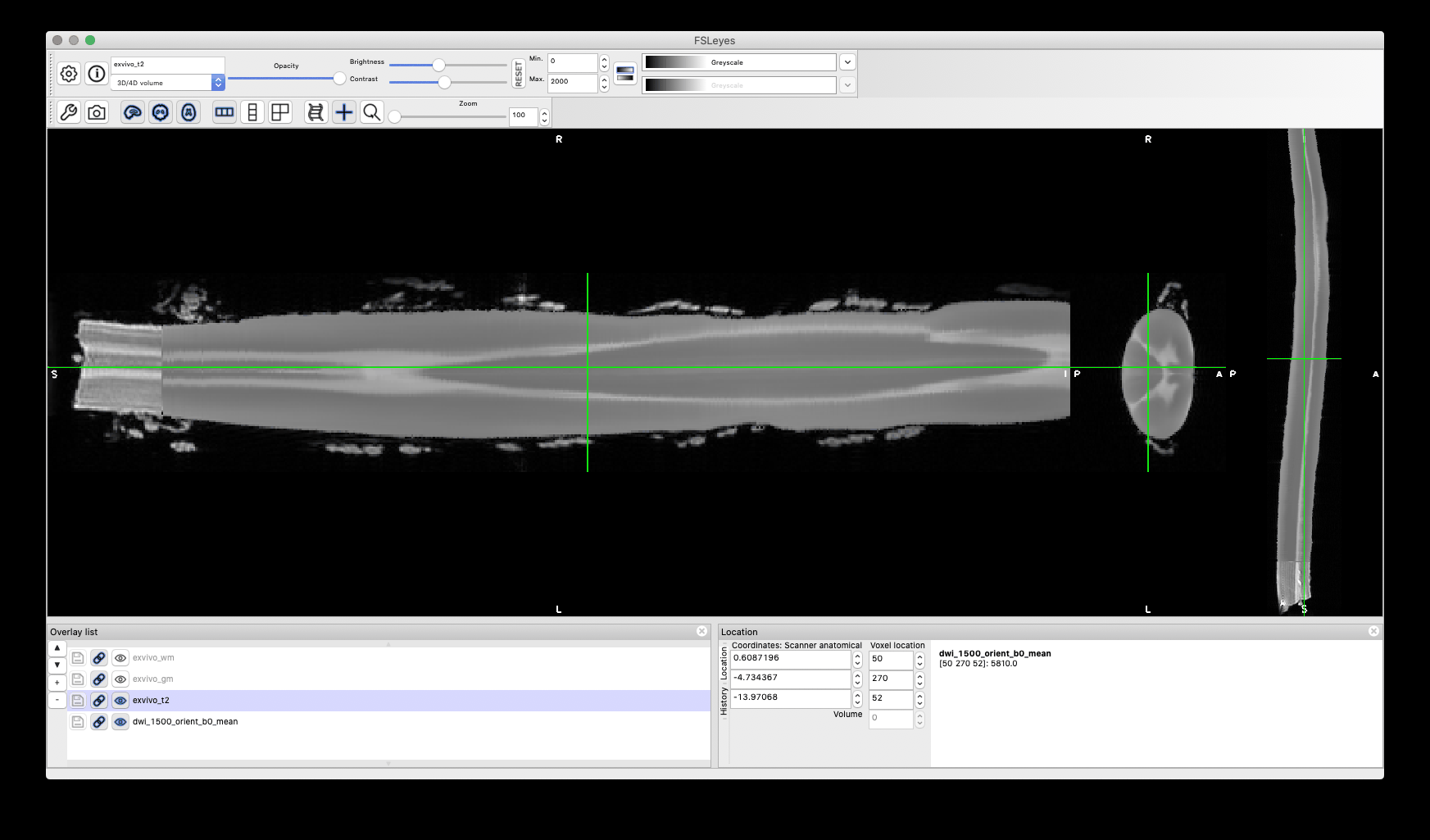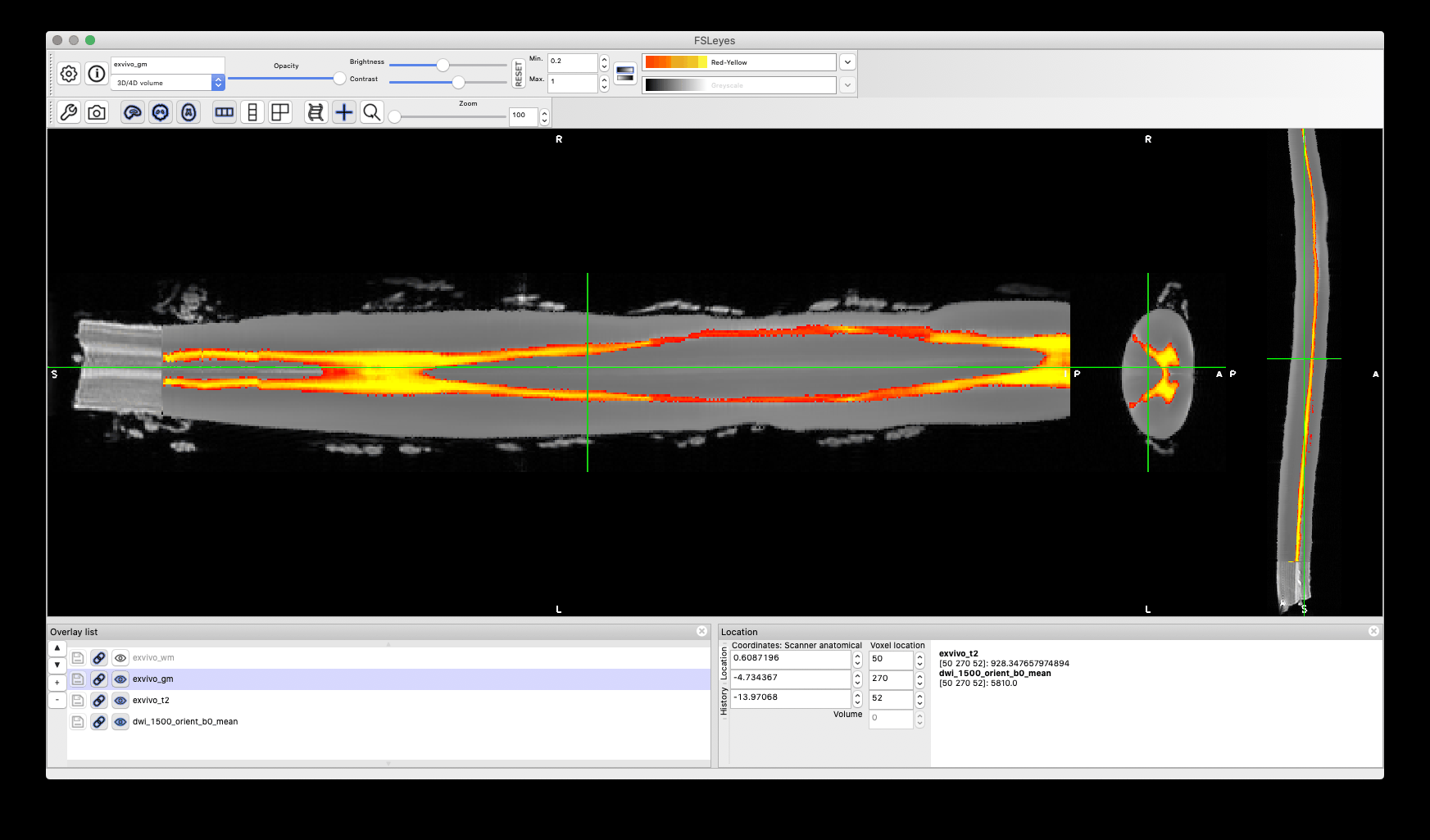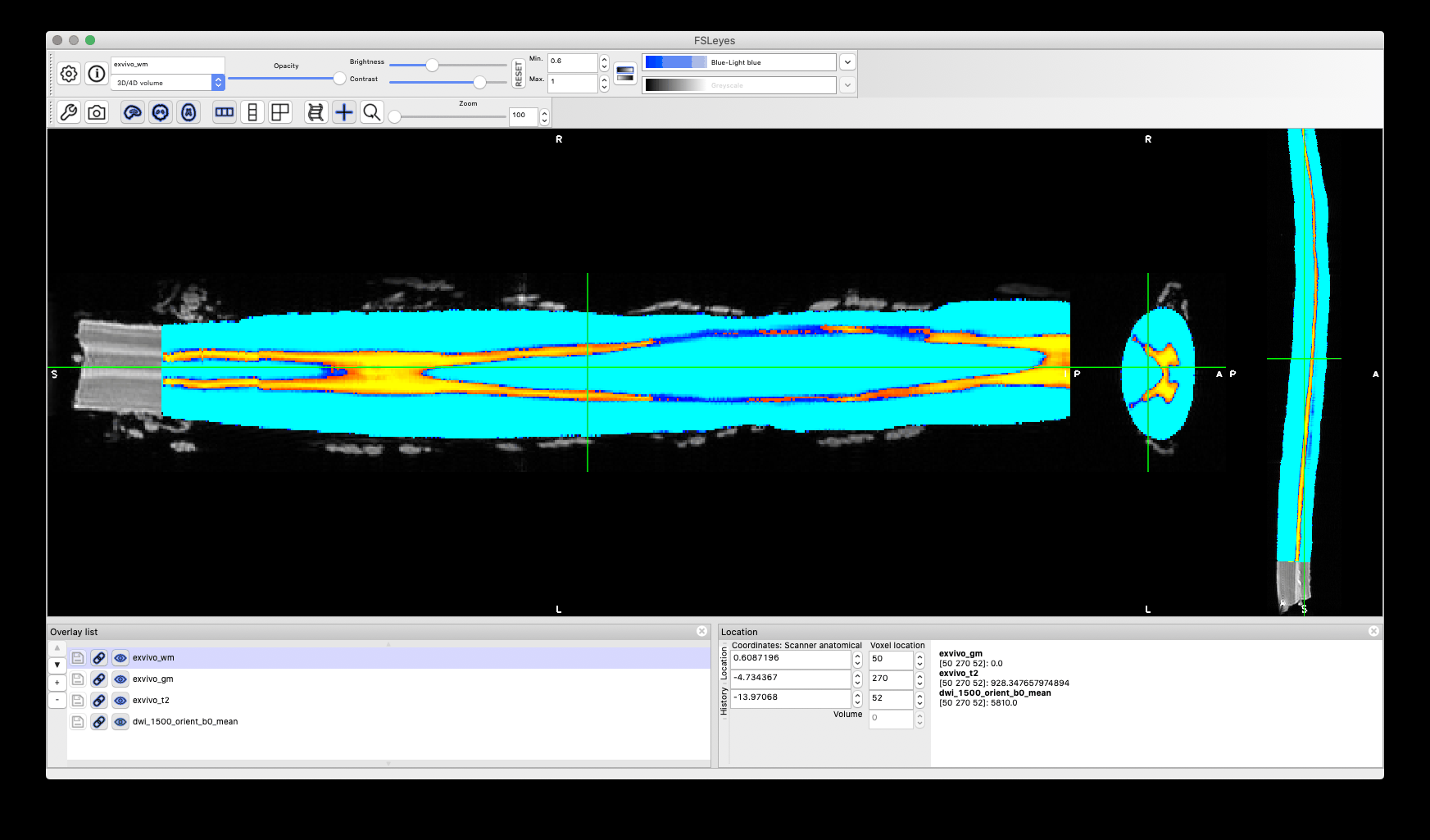
Hi SCT Team,
I’m trying to register my high resolution Ex vivo human dwi data into T1 template here https://github.com/sct-pipeline/exvivo-template.
I did the following steps but ended up with an error in template registration…
1-Dicom data imported from Bruker animal MRI system. Converted to nifty using mrconvert as package from MRtrix dcm to nifty and also used dcm2niix software for comparison…
The fslhd output using dcm2niix converter is below
filename dwi_b0_b1500_reordered.nii.gz
sizeof_hdr 348
data_type FLOAT32
dim0 4
dim1 100
dim2 512
dim3 100
dim4 20
dim5 1
dim6 1
dim7 1
vox_units mm
time_units s
datatype 16
nbyper 4
bitpix 32
pixdim0 -1.000000
pixdim1 0.200000
pixdim2 0.156250
pixdim3 0.200000
pixdim4 0.300000
pixdim5 0.000000
pixdim6 0.000000
pixdim7 0.000000
vox_offset 352
cal_max 0.000000
cal_min 0.000000
scl_slope 1.000000
scl_inter 0.000000
phase_dim 0
freq_dim 0
slice_dim 0
slice_name Unknown
slice_code 0
slice_start 0
slice_end 0
slice_duration 0.000000
toffset 0.000000
intent Unknown
intent_code 0
intent_name
intent_p1 0.000000
intent_p2 0.000000
intent_p3 0.000000
qform_name Scanner Anat
qform_code 1
qto_xyz:1 0.000000 0.000000 0.200000 -9.800000
qto_xyz:2 -0.200000 0.000000 -0.000000 3.000000
qto_xyz:3 0.000000 0.156250 -0.000000 -39.843750
qto_xyz:4 0.000000 0.000000 0.000000 1.000000
qform_xorient Anterior-to-Posterior
qform_yorient Inferior-to-Superior
qform_zorient Left-to-Right
sform_name Scanner Anat
sform_code 1
sto_xyz:1 0.000000 0.000000 0.200000 -9.800000
sto_xyz:2 -0.200000 0.000000 -0.000000 3.000000
sto_xyz:3 0.000000 0.156250 0.000000 -39.843750
sto_xyz:4 0.000000 0.000000 0.000000 1.000000
sform_xorient Anterior-to-Posterior
sform_yorient Inferior-to-Superior
sform_zorient Left-to-Right
file_type NIFTI-1+
file_code 1
descrip 6.0.4:ddd0a010
The fslhd output using ***mrconvert*** converter is below
filename dwi_1500.nii.gz
sizeof_hdr 348
data_type FLOAT32
dim0 4
dim1 100
dim2 512
dim3 100
dim4 20
dim5 1
dim6 1
dim7 1
vox_units mm
time_units s
datatype 16
nbyper 4
bitpix 32
pixdim0 1.000000
pixdim1 0.156250
pixdim2 0.200000
pixdim3 0.200000
pixdim4 nan
pixdim5 0.000000
pixdim6 0.000000
pixdim7 0.000000
vox_offset 352
cal_max 0.000000
cal_min 0.000000
scl_slope 1.000000
scl_inter 0.000000
phase_dim 0
freq_dim 0
slice_dim 0
slice_name Unknown
slice_code 0
slice_start 0
slice_end 0
slice_duration 0.000000
toffset 0.000000
intent Unknown
intent_code 0
intent_name
intent_p1 0.000000
intent_p2 0.000000
intent_p3 0.000000
qform_name Scanner Anat
qform_code 1
qto_xyz:1 0.000000 0.000000 0.200000 -9.800000
qto_xyz:2 -0.156250 0.000000 0.000000 3.000000
qto_xyz:3 0.000000 -0.200000 0.000000 40.000000
qto_xyz:4 0.000000 0.000000 0.000000 1.000000
qform_xorient Anterior-to-Posterior
qform_yorient Superior-to-Inferior
qform_zorient Left-to-Right
sform_name Scanner Anat
sform_code 1
sto_xyz:1 0.000000 0.000000 0.200000 -9.800000
sto_xyz:2 -0.156250 0.000000 0.000000 3.000000
sto_xyz:3 0.000000 -0.200000 0.000000 40.000000
sto_xyz:4 0.000000 0.000000 0.000000 1.000000
sform_xorient Anterior-to-Posterior
sform_yorient Superior-to-Inferior
sform_zorient Left-to-Right
file_type NIFTI-1+
file_code 1
descrip 6.0.4:ddd0a010
I provided these information to make sure which software should be used to handle the post-processing in correct way when using SCT. Please note, I used dcm2niix nifty output for first registration and mrconvert output for second attempt. However, I got the same error…
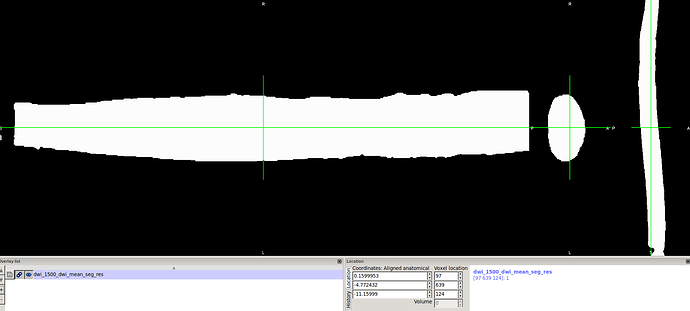
**dwi mean was computed and cord segmentation produced using sct_propseg
3- dwi date splitted using sct_image flag -split t to get b0 image
4-b0 and cord seg were resampled to T1 template which is 0.08mm3
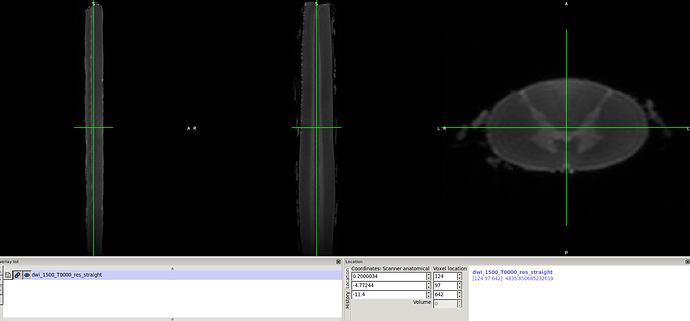
5-sct_straighten_spinalcord -i b0_resambeled.nii.gz -s cord_segmentation_resampled.nii.gz used to straighten the cord.
result..
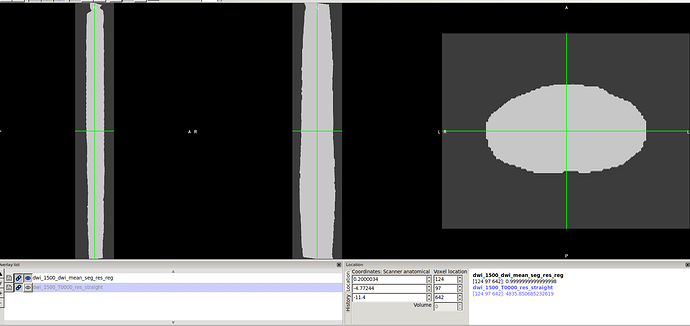
6-I apply the transformation out of b0 in the previous step to the cord segmentation as well using sct_apply_transfo command.
7-Now, I tried to create the segments labels using the following syntax along Z direction…
sct_label_utils -i dwi_1500_T0000_res_straight.nii.gz -create 125,935,125,3:125,311,125,4:125,603,125,5:125,392,125,6:125,222,125,7 -o sc_labels.nii.gz
Terminal output error…
Traceback (most recent call last):
File "/data/home/s4333238/.ssh/spinalcordtoolbox/spinalcordtoolbox/scripts/sct_label_utils.py", line 413, in <module>
main(sys.argv[1:])
File "/data/home/s4333238/.ssh/spinalcordtoolbox/spinalcordtoolbox/scripts/sct_label_utils.py", line 261, in main
out = sct_labels.create_labels_empty(img, labels)
File "/data/home/s4333238/.ssh/spinalcordtoolbox/spinalcordtoolbox/labels.py", line 53, in create_labels_empty
out = _add_labels(zeros_like(img), coordinates)
File "/data/home/s4333238/.ssh/spinalcordtoolbox/spinalcordtoolbox/labels.py", line 84, in _add_labels
img.data[int(x), int(y), int(z)] = v
**IndexError: index 935 is out of bounds for axis 1 with size 195**
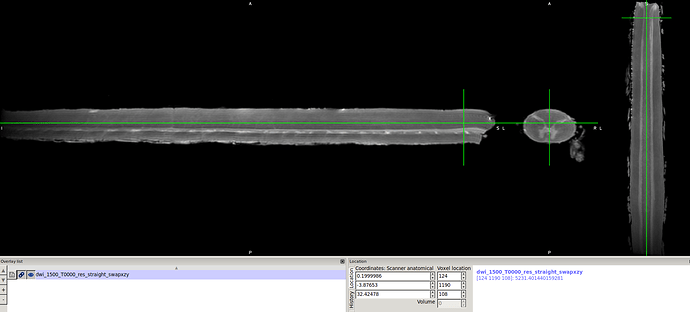
So, I tried to swapdim to get the labels correctly pointed in the right axis I used the following syntax
fslswapdim dwi_res_straight.nii.gz x z y dwi_res_straight_xzy.nii.gz
Then I re-ran the syntax using dwi_res_straight_xzy.nii.gz as input…
output…
Generating output files...
Done! To view results, type:
fsleyes dwi_1500_T0000_res_straight_swapxzy.nii.gz sc_labels.nii.gz &
8-After I had the data and cord seg. resampled,straightened,labeled, I ran the following command for registration…
sct_register_to_template -i dwi_stra_xzy.nii.gz -s dwi_mean_seg_res_reg_xzy.nii.gz -lspinal sc_labels.nii.gz -t exvivo-template/ -c t1
Output…ERROR
Check template files...
OK: exvivo-template/template/template.nii.gz
OK: exvivo-template/template/label_spinalsegments.nii.gz
OK: exvivo-template/template/mask_spinalcord.nii.gz
Check parameters:
Data: dwi_stra_xzy.nii.gz
Landmarks: sc_labels.nii.gz
Segmentation: dwi_mean_seg_res_reg_xzy.nii.gz
Path template: exvivo-template/
Remove temp files: 1
Check input labels...
Creating temporary folder (/tmp/sct-20210306153353.274717-register_to_template-3ql8kvwm)
Copying input data to tmp folder and convert to nii...
Check if provided labels are available in the template
To avoid intensity overflow due to convertion to +uint8+, intensity will be rescaled to the maximum quantization scale
Binarize segmentation
Resample data to 1mm isotropic...
load data...
Done! To view results, type:
fsleyes data_1mm.nii &
load data...
Done! To view results, type:
fsleyes seg_bin_1mm.nii.gz &
To avoid intensity overflow due to convertion to +uint8+, intensity will be rescaled to the maximum quantization scale
Change orientation of input images to RPI...
Straighten the spinal cord using centerline/segmentation...
Creating temporary folder (/tmp/sct-20210306153822.455291-straighten_spinalcord-p1h_7a5e)
Copy files to tmp folder...
**Image /tmp/sct-20210306153353.274717-register_to_template-3ql8kvwm/template_seg.nii.gz has different qform and sform matrices. This can produce incorrect results. Consider setting the two matrices to be equal by running:**
sct_image -i /tmp/sct-20210306153353.274717-register_to_template-3ql8kvwm/template_seg.nii.gz -set-sform-to-qform -o /tmp/sct-20210306153353_same-affine.274717-register_to_template-3ql8kvwm/template_seg.nii.gz.
Traceback (most recent call last):
File "/data/home/s4333238/.ssh/spinalcordtoolbox/spinalcordtoolbox/scripts/sct_register_to_template.py", line 1281, in <module>
main(sys.argv[1:])
File "/data/home/s4333238/.ssh/spinalcordtoolbox/spinalcordtoolbox/scripts/sct_register_to_template.py", line 533, in main
sc_straight.straighten()
File "/data/home/s4333238/.ssh/spinalcordtoolbox/spinalcordtoolbox/straightening.py", line 111, in straighten
Image(self.centerline_reference_filename, check_sform=True).save(os.path.join(path_tmp, "centerline_ref.nii.gz"))
File "/data/home/s4333238/.ssh/spinalcordtoolbox/spinalcordtoolbox/image.py", line 311, in __init__
raise ValueError("Image sform does not match qform")
**ValueError: Image sform does not match qform**
Then I ran the following syntax to set sform and qform on my input data,seg and labels.
sct_image data.nii.gz -set-sform-to-qform data_corrected.nii.gz
After I’ve done it also for other inputs labels and segmentation, I re-ran the registration on the corrected one.
The final data used for registration after processing/ Step 1 I used dcm2niix output for post-processing…
filename dwi_T0000_straight_resa_xzy.nii.gz
sizeof_hdr 348
data_type FLOAT64
dim0 3
dim1 248
dim2 1004
dim3 248
dim4 1
dim5 1
dim6 1
dim7 1
vox_units mm
time_units s
datatype 64
nbyper 8
bitpix 64
pixdim0 -1.000000
pixdim1 0.079839
pixdim2 0.079993
pixdim3 0.079839
pixdim4 1.000000
pixdim5 1.000000
pixdim6 1.000000
pixdim7 1.000000
vox_offset 352
cal_max 0.000000
cal_min 0.000000
scl_slope 1.000000
scl_inter 0.000000
phase_dim 0
freq_dim 0
slice_dim 0
slice_name Unknown
slice_code 0
slice_start 0
slice_end 0
slice_duration 0.000000
toffset 0.000000
intent Unknown
intent_code 0
intent_name
intent_p1 0.000000
intent_p2 0.000000
intent_p3 0.000000
qform_name Unknown
qform_code 0
qto_xyz:1 0.079839 -0.000000 -0.000000 -9.720161
qto_xyz:2 0.000000 0.000000 0.079839 -16.800001
qto_xyz:3 0.000000 0.079993 -0.000000 -40.468750
qto_xyz:4 0.000000 0.000000 0.000000 1.000000
qform_xorient Left-to-Right
qform_yorient Inferior-to-Superior
qform_zorient Posterior-to-Anterior
sform_name Unknown
sform_code 2
sto_xyz:1 0.079839 0.000000 0.000000 -9.720161
sto_xyz:2 0.000000 0.000000 0.079839 -16.800001
sto_xyz:3 0.000000 0.079993 0.000000 -40.468750
sto_xyz:4 0.000000 0.000000 0.000000 1.000000
sform_xorient Left-to-Right
sform_yorient Inferior-to-Superior
sform_zorient Posterior-to-Anterior
file_type NIFTI-1+
file_code 1
descrip 6.0.4:ddd0a010
**The final data used for registration after processing/ Step 2, I used mrconvert output for post-processing..**
filename dwi_T0000_res_xzy.nii.gz
sizeof_hdr 348
data_type FLOAT64
dim0 3
dim1 249
dim2 1286
dim3 195
dim4 1
dim5 1
dim6 1
dim7 1
vox_units mm
time_units s
datatype 64
nbyper 8
bitpix 64
pixdim0 -1.000000
pixdim1 0.080000
pixdim2 0.080000
pixdim3 0.080128
pixdim4 0.000000
pixdim5 0.000000
pixdim6 0.000000
pixdim7 0.000000
vox_offset 352
cal_max 0.000000
cal_min 0.000000
scl_slope 1.000000
scl_inter 0.000000
phase_dim 0
freq_dim 0
slice_dim 0
slice_name Unknown
slice_code 0
slice_start 0
slice_end 0
slice_duration 0.000000
toffset 0.000000
intent Unknown
intent_code 0
intent_name
intent_p1 0.000000
intent_p2 0.000000
intent_p3 0.000000
qform_name Scanner Anat
qform_code 1
qto_xyz:1 0.080000 -0.000000 -0.000000 -9.720001
qto_xyz:2 0.000000 0.000000 0.080128 -12.544872
qto_xyz:3 0.000000 0.080000 -0.000000 -62.799995
qto_xyz:4 0.000000 0.000000 0.000000 1.000000
qform_xorient Left-to-Right
qform_yorient Inferior-to-Superior
qform_zorient Posterior-to-Anterior
sform_name Scanner Anat
sform_code 0
sto_xyz:1 0.000000 0.000000 0.000000 0.000000
sto_xyz:2 0.000000 0.000000 0.000000 0.000000
sto_xyz:3 0.000000 0.000000 0.000000 0.000000
sto_xyz:4 0.000000 0.000000 0.000000 1.000000
sform_xorient Unknown
sform_yorient Unknown
sform_zorient Unknown
file_type NIFTI-1+
file_code 1
descrip 6.0.4:ddd0a010
I ended up with the same error… I wonder whether mty steps were in right track or not. Is there any comment regarding the data header info. as I’m really confused about which oriention is recommended.
Sorry for the long post but I tried to provide you with more details as possible to track the error and for others if they get same issue in future…
Many thanks in advance…
Cheers,
Ibrahim