-
A description of the problem.
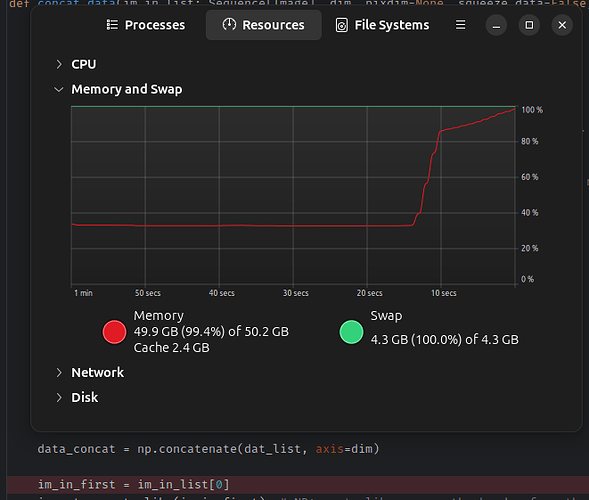
I have a functional file in the subject space that I want to transform into PAM50 standard space. I’ve also acquired a warping field using the Tmean image of the functional file. When I run the transform, SCT “splits the image along T dimension” and transforms the 3D image of each time point, but gets stuck at the step “Merge file back…”. -
Commands and terminal output. (What are the exact commands you used? Please copy and paste the full output of the command from your terminal.)
sct_apply_transfo -i /Users/wgh3051/Desktop/SCVR_Pilot01_ses02/1/moco/SCVR_Pilot01_ses02_SC_REST_30_25_2000_1x1x3mm_ZOOMit_20240422140801_1_trnc_moco.nii.gz -d /Users/wgh3051/sct_6.5/data/PAM50/template/PAM50_t2s.nii.gz -w /Users/wgh3051/Desktop/SCVR_Pilot01_ses02/1/registration_output/warp_func_mean2PAM50_t2s.nii.gz -o /Users/wgh3051/Desktop/RampUp-ICA/SCVR_Pilot01_ses02_1_PAM50.nii.gz
- System information.
--
Spinal Cord Toolbox (6.5)
sct_check_dependencies
--
SYSTEM INFORMATION
------------------
SCT info:
- version: 6.5
- path: /Users/wgh3051/sct_6.5
OS: osx (macOS-10.16-x86_64-i386-64bit)
CPU cores: Available: 11, Used by ITK functions: 11
RAM: Total: 18432MB, Used: 1919MB, Available: 1442MB
OPTIONAL DEPENDENCIES
---------------------
Check FSLeyes version...............................[OK] (1.10.0)
MANDATORY DEPENDENCIES
----------------------
Check Python executable.............................[OK]
Using bundled python 3.9.20 (main, Oct 3 2024, 02:27:54)
[Clang 14.0.6 ] at /Users/wgh3051/sct_6.5/python/envs/venv_sct/bin/python
Check if acvl_utils is installed....................[OK]
Check if dipy is installed..........................[OK] (1.8.0)
Check if ivadomed is installed......................[OK] (2.9.10)
Check if matplotlib is installed....................[OK] (3.9.3)
Check if matplotlib-inline is installed.............[OK]
Check if monai is installed.........................[OK] (1.4.0)
Check if nibabel is installed.......................[OK] (5.3.2)
Check if nilearn is installed.......................[OK] (0.10.4)
Check if nnunetv2 is installed......................[OK]
Check if numpy is installed.........................[OK] (1.26.4)
Check if onnxruntime is installed...................[OK] (1.19.2)
Check if pandas is installed........................[OK] (1.5.3)
Check if portalocker is installed...................[OK] (3.0.0)
Check if psutil is installed........................[OK] (6.1.0)
Check if pyqt5 (5.12.3) is installed................[OK] (5.12.3)
Check if pyqt5-sip is installed.....................[OK]
Check if pystrum is installed.......................[OK] (0.4)
Check if pytest is installed........................[OK] (8.3.4)
Check if pytest-cov is installed....................[OK] (6.0.0)
Check if requests is installed......................[OK] (2.32.3)
Check if requirements-parser is installed...........[OK] (0.11.0)
Check if scipy is installed.........................[OK] (1.13.1)
Check if scikit-image is installed..................[OK] (0.24.0)
Check if scikit-learn is installed..................[OK] (1.5.2)
Check if totalspineseg is installed.................[OK]
Check if xlwt is installed..........................[OK] (1.3.0)
Check if tqdm is installed..........................[OK] (4.67.1)
Check if transforms3d is installed..................[OK] (0.4.2)
Check if urllib3 is installed.......................[OK] (2.2.3)
Check if pytest_console_scripts is installed........[OK]
Check if pyyaml is installed........................[OK] (6.0.2)
Check if voxelmorph is installed....................[OK] (0.2)
Check if wquantiles is installed....................[OK] (0.4)
Check if xlsxwriter is installed....................[OK] (3.2.0)
Check if spinalcordtoolbox is installed.............[OK]
Check ANTs compatibility with OS ...................[OK]
Check PropSeg compatibility with OS ................[OK]
Check if figure can be opened with PyQt.............[OK]
Check if figure can be opened with matplotlib.......[OK] (Using GUI backend: 'qtagg')
Check data dependency 'PAM50'.......................[OK]
Check data dependency 'deepseg_gm_models'...........[OK]
Check data dependency 'deepseg_sc_models'...........[OK]
Check data dependency 'deepseg_lesion_models'.......[OK]
Check data dependency 'deepreg_models'..............[OK]
Check data dependency 'PAM50_normalized_metrics'....[OK]
Check data dependency 'binaries_osx'................[OK]
- File upload.
https://drive.google.com/drive/folders/1iOilFp50bL2me41T8-QVtqPohpHNk07K?usp=sharing