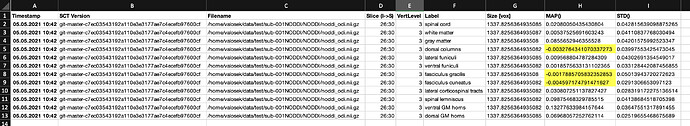
When I run sct_extract_metric using map method on orientation dispersion index (ODI) metric (extracted using NODDI) I get negative values for some labels (tracts):
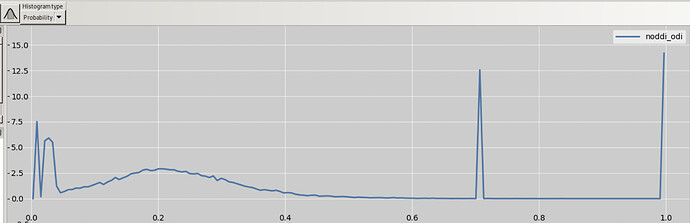
ODI image itself contains values only in interval 0-1, though:
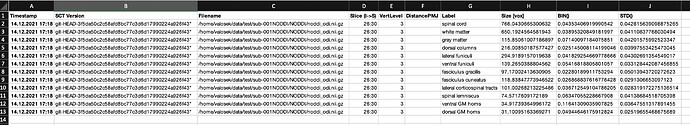
If I use method bin, extracted values are reasonable (non-negative):
I am using the latest SCT version (v5.4). Command looks like:
sct_extract_metric -i noddi_odi.nii.gz -f label/atlas -method map -o noddi_odi_C3_map_method.csv -vert 3 -l 50,51,52,53,54,55,56,57,58,59,61,62 -v 1 -append 1
I can provide the data in private message.
Hi Jan,
The MAP method is based on the assumptions that:
- each region is relatively homogeneous in terms of ‘true’ signal;
- the amount of noise is reasonable;
- the number of voxels used to compute the inference is sufficient;
There are relevant details (with links to papers and simulations) in this link.
In your data, I can see that signal in the WM is close to zero (~0.005), so it is not surprising that, with a bit of noise, some tracts within the WM would be estimated below zero.
Now, when comparing these results to the BIN results, even though the BIN results are never negative (they cannot, as you show in the histogram), it does not imply that the BIN results are “more valid”. They could still be way off the true value because of partial volume effect.
To find out about the validity of the MAP method in your case, I would look at a few things. First, how homogeneous is the signal in the surrounding CSF? I’m asking this because the MAP uses priors from the mean GM, WM and CSF signal to regularize values. So, if the CSF signal is salt and pepper, then it will likely not be stable enough to use it as a prior, and in this case you should rather use the ML method, or maybe the WA (or BIN).
Hi Julien,
Thank you for your answer and sorry for my delayed response.
In your data, I can see that signal in the WM is close to zero (~0.005), so it is not surprising that, with a bit of noise, some tracts within the WM would be estimated below zero.
Generally, orientation dispersion index (ODI) values are “low”. GM has values in interval 0.05-0.15 and WM even lower (0.001-0.05). See for example Fig 8 in Schilling et al., 2019 or Fig 3 in Grussu et al, 2015.
To find out about the validity of the MAP method in your case, I would look at a few things. First, how homogeneous is the signal in the surrounding CSF?
Compared with the WM and GM values, ODI in CSF is much higher. In case of my data, CSF has ODI=0.7. Moreover, this value is almost same across all voxels in CSF.
I’m asking this because the MAP uses priors from the mean GM, WM and CSF signal to regularize values.
Since CSF values are kind of “weird” in my data, I have a hunch that this can be the source of error during the MAP estimation.
I tried the median method and got similar values for WM and GM as reported in Table 1 in Grussu et al, 2015.
So, thank you one more time for your answer. I have to further explore fitting of NODDI on my data (particularly in the CSF).