Hi SCT Forum,
How many samples are represented are the “MAP()” and “STD()” columns in the output of sct_extract_metric (attached). We would like to combine outputs and perform statistical tests on different levels and segments, which often require the data sample size (i.e., number of voxels on which the data is based).
Based on other forum questions, the “Size [vox]” column is voxel fraction, which explains why it is not an integer. And since the value is the same for all segments, it cannot be the number of samples used to calculate the metric value.
Thanks in advance for any help and education!
FA_06.csv (6.8 KB)
Hi @Bryan_Zheng,
Thank you for reaching out. As you rightfully guessed, the number of voxels used to extract the metric for the specific label is indicated under the column “Size [vox]”. However, I see that you are using an old version of SCT (v4.3), where there was a bug in the display of the proper size per tract when using the ml and map methods. You have two options:
-
You can run sct_extract_metric with -method wa, which should give you a different number of voxels across the different tracts.
-
You can install the latest version of SCT (I recommend that option).
Cheers,
Julien
@jcohenadad
Thanks for the quick solution! We’ve updated to SCT v5.4 and changed the -method from map to wa. Now we do get different sizes per tract! Seeing as the values are not integers (voxel fractions?: https://doi.org/10.1016/j.neuroimage.2016.10.009), how does one obtain the “sample size” for the average? For example, the attached result is from one DWI slice (z = 3). We want to combine the statistical measures for levels with a lesion and compare against normal levels, which requires the sample size.
FA_z3.csv (7.8 KB)
Hi,
Firstly, you did not need to update to v5.4 and change for -method wa. My suggestions were (possibly) mutually exclusive. With v5.4 you should be table to keep map while still seing the size per tract. But it’s up to you what method you opt for, as long as it is properly justified from a methodological standpoint (each method has its own assumptions and limitations).
Now regarding your question about sample size: It is difficult for me to advise on how you should do your statistics if I don’t have the “full picture” of your experimental setup (e.g., what is the null hypothesis, what are the assumptions, heteroscedasticity, etc.). From what I understand, you would like to compare the average FA metrics between one “pathological” region and one “healthy” region, but in this case, the “sample” you are referring to should not be the number of voxels, given the high spatial autocorrelations in the data (ie: the assumption of sample independence is violated). You should ideally do your statistics by comparing average values across participants, and use the number of participants in each group as the sample size.
That being said, there are solutions to compute statistics with non-integer degrees of freedom. See for example (source here):

Hey @jcohenadad, thanks for the continued help. For some reason when I first ran the analysis using the updated SCT it still returned uniform values, though the behavior is now as expected upon double checking - sorry for the confusion.
With regards to the statistical comparisons, your point about non-independence is well taken and we will still look into analyzing possible non-integer degrees of freedom. Thanks! We do plan to use population data for studies in general, though in this particular case, the analysis is more of a “case report” style where we would like to compare slices affected by a lesion to levels not. Perhaps the correct sample size here is then the number of slices used in each group (only 15 slices total, with ~3 affected by the lesion)? In any case, it does seem like the comparison may still be very underpowered.
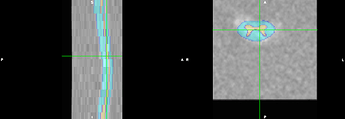
I understand the technical issue is now resolved so any further guidance is tangential. But the basic goal is to give our best guess as to what type of lesion is present (working hypothesis is demyelination, where the clinical data is inconclusive) using DTI metrics. We also have MTR data on this patient, though its relevance is even less clear. On inspection, there does appear to be some broad changes in metrics, for example FA (attached, with some supporting screenshots to just confirm data quality and purely clinical post-gado imaging demonstrating contrast enhancement).
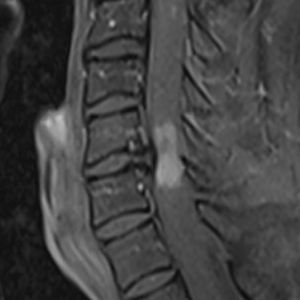
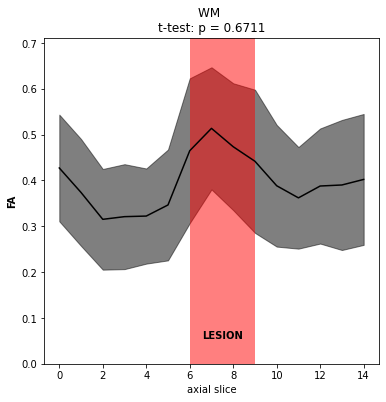
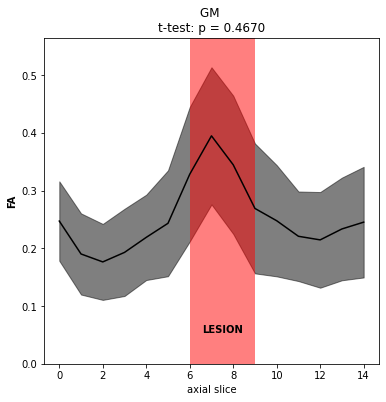
Let us know if you have any thoughts and questions on the “fuller picture”, though as stated above, your response up to this point has been great and more than enough!
I see. In this case I would indeed suggest to consider one slice as a sample. One possibility would be to compute the confidence interval of the FA for the non-lesioned slices, and test if the FA in the lesioned slices fall outside that confidence interval. But I am not a statistician expert, so please don’t quote me on this