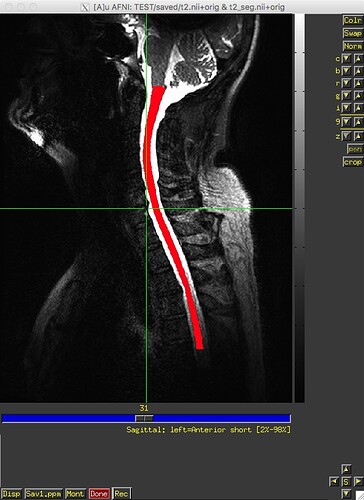
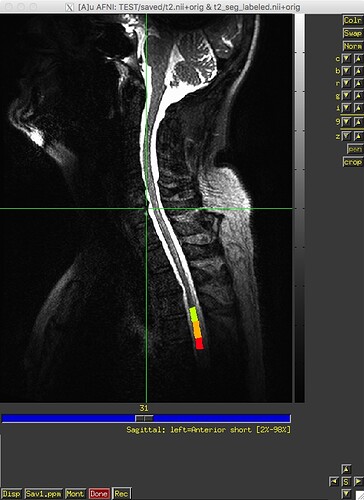
Hi developers:
Sorry for yet another post, but…
I ran sct_label_vertebrae from release 3.2.7 on 12 datasets last week, and, as far as I can recall, it worked fine on all datasets. I recently downloaded v4.0.0-beta.2 and re-ran sct_label_vertebrae on the same 12 datasets – and it failed on one dataset. The text and image outputs from sct_deepseg_sc and sct_label_vertebrae from this one subject are attached. Any insight into what’s going on would be greatly appreciated. Thank you.
-Rob
SCT_20190418_ver_4_0_0_beta_2.txt (5.1 KB)
Hi Rob,
Some changes are indeed expected. We noticed few glitches with v3.2.7 on the spineGeneric data, so we made sct_label_vertebrae more robust to these datasets. It is possible that, by making it more robust to these datasets, it became less robust to other types of data.
If you can send us the problematic data, we can see if there is something quick we can try to make 4.0.0 work better on your data.
Cheers,
Julien
1 Like
Thanks, Julien.
I just shared this problematic dataset with you.
-Rob
Hi Rob,
I confirm that, for the dataset you sent me, automatic labeling works with 3.2.7 but not with 4.0.0-beta.2.
If this only happens in ~10% of the cases, I suggest you do a manual initialization using flag:
sct_label_utils -i t2.nii -create-viewer 3 -o labelc2c3.nii.gz
sct_label_vertebrae -i t2.nii -s t2_seg.nii -c t2 -initlabel labelc2c3.nii.gz -ofolder initlabel -qc ~/qc-sct
An example workflow would consist in running vertebral labeling for all subjects, then check the qc (notice the flag -qc), then for the problematic subjects generate a manual label, then re-run your analysis by pointing to the label file if it exists. That way you ensure a fully reproducible pipeline, allowing manual intervention where necessary.
That being said, if you find that automatic labeling fails in >25% of the case, we can easily generate a new model specific to your data, to improve robustness.
Cheers,
Julien
1 Like
Hi Julien,
Thank you for verifying the differences between 3.2.7 and 4.0.0-beta.2 for this dataset. And I appreciate the suggested workflow - I will modify my code accordingly.
Cheers,
-Rob
Hi Rob,
I just noticed a bug in v4.0.0-beta.2, which made sct_label_vertebrae under-perform. This will be fixed in the next release. You can already test the fix in this branch.
Cheers,
Julien
1 Like
Hi Julien,
Thanks for the update. A bug likely explains why the labeling for this one challenging data set worked with 3.2.7 but failed with 4.0.0-beta.2. I will see if I can test this fix, but the installations of 4.0.0-beta2 on macOS 10.14.x are unfortunately still not working (my last git pull & test of the master branch was two days ago).
Cheers,
-Rob
Hi Rob,
I was talking about another branch (not merged with the master branch yet).
To test this branch, you can do:
git checkout -b jca/py3k origin/jca/py3k
./install_sct
However, if you prefer, you can also wait for it to be merged with master.
Cheers,
Julien
1 Like
Hi Julien,
I imagine that you are busy with the ISMRM but I still wanted to let you know that sct_label_vertebrae shows a different behavior since the update. As far as I understand, the branch that you mentioned before in this thread was merged with the master branch, so what I observe might be something different.
The behavior that I observe is the following:
- the labeling is more robust and works in more subjects than before
- in many cases, the disc c2/c3 is identified correctely
- nevertheless, the hight of other discs/vertrebrae is less precise than before (in several subjects)
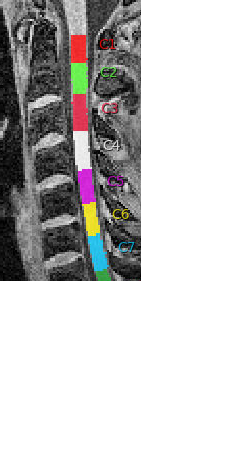
I am focused on vertebrae C5, so my analysis focuses on optimizing the normalization of C5 to the template. That worked nicely before but now the hight of my normalized T1 images is less precise because the labeling of C5 is also less precise. To improve results, it helps to label c4/c5 manually but that means I would need to do that for many subjects.
If there is no quick fix for this, can I just download the function from an older version?
Best,
Alexandra
Hi Alexandra,
this is weird. The discs are very well visible. What version are you running and what syntax did you type?
Hi Julien,
I updated to version 4.0.0-beta.4 recently.
I call the function like this:
sct_label_vertebrae -i t1.nii -s t1_seg.nii -c t1 -qc ~/qc_multiSubj
And I found the labeling results from the same subject in an older version where it looks perfect:
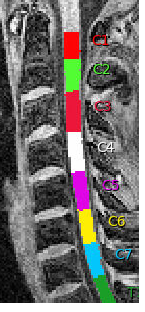
The qc results in the browser don’t display the version number, so I don’t know for sure which version that was but it outputs this:
git-master-5de003c7962051320e27c2ba579bf9f5bcc73a39
I don’t know how to relate this number to a version but maybe it tells you something…
Best,
Alexandra
Would you mind sharing the t1 and t1_seg with me so I can see what’s happening?
Hi @Alexandra_Tinnermann,
I was able to track down the source of the performance loss. A ticket has been opened here and will be addressed with high priority.
Thank you very much for letting us know!
Julien
Hi @Alexandra_Tinnermann,
The issue has been solved in the latest release (4.0.0-beta.5).
Let us know how it goes,
Best,
Julien
Hi Julien,
with the new release, everything looks as good as before!
Thanks a lot for all your effort!
Best,
Alexandra