Hi all,
I am trying to register a diffusion-weighted sequence to T2 (sagittal) sequence in order to apply a spinal tract atlas (PAM50). Here is the command of interest:
sct_register_multimodal -i ../sag_t2/sag_t2_seg.nii -d dmri_mean.nii.gz -identity 1 -x nn -param step=1,algo=slicewise
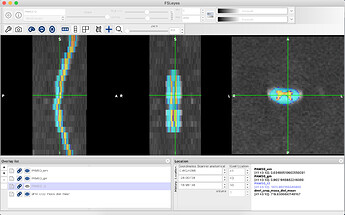
Unfortunately, the geometry of the diffusion data seems to be sheared such that any registration ends up looking like this (DWI is overlayed on segmented spinal cord from sagittal T2 image):
Is there any way to register these sequences such that the registration is valid throughout the entire cord?
Note: I ultimately want to calculate metrics (FA, MD) for specific spinal tracts at specific levels. Right now I can only extract aggregate data throughout the entire cord, where the data is invalid anyways because the registration is off. So if possible, a workaround that may be easier would be to extract individual levels (C1-C2, C2-C3, …, etc.) and register them individually with the anatomical (T2) data. If anyone thinks that is possible and preferable, please let me know!
Thanks! 1st forum post - I’m really impressed with SCT so far. Let me know if any additional information would be helpful!