Hi,
I’m trying to register mFFE 3D volume consisting of 14 slices to the template space. I’ve registered several scans already, but for some of them the registration is rotated, misaligned, not registered, or only has one slice that is registered. I’ve attached a ppt showing the issues. I’m particularly concerned about the scans that are not registering or have compressed registration. These scans have mFFE crops that work and remain in the same space, are segmented appropriately, and have correct labels, but don’t register to the template space. When analyzing the deformation fields in mipav, the field is completely dark and the transformation is not apparent. The deformation fields for the empty registered scans are all very large (~3.48*10^33). I’m using the following command for registration: sct_register_to_template -i mFFE_crop.nii -s mFFE_crop_seg.nii -l labels_vert.nii.gz -c t2 -param step=1,type=seg,algo=centermassrot,metric=MeanSquares:step=2,type=seg,algo=columnwise,metric=MeanSquares -ref subject. Vertebral straightening by removing “-ref subject” does not change the issue. Please let me know if you have any suggestions.
Faulty Registrations copy.pptx (607.4 KB)
Thanks,
Ashwin
Hi Ashwin,
It looks like the rotation estimation is not robust in your case, so I would remove it (replacing centermassrot by centermass). Also, for some cases where the registration gives you a blank image, I would make sure the segmentation and/or the vertebral labeling are correct. I recommend you use the -qc flag for your segmentation and labeling tasks, they enable quick QC to make sure these steps ran OK.
If these tips don’t help, please don’t hesitate to send me 1-2 representative cases and I will see what I can do.
Cheers,
Julien
Hi Dr. Cohenadad,
Thanks for your response. The centermass suggestion appears to fix the observed rotation and misalignment. However, the empty or compressed scans still have an empty anat2template.nii, but after adding the -qc flag, it appears that there is a registered image when looking at the index.html. I manually labeled the vertebrae, so vertebral labeling should be correct. I have uploaded the folder with the empty scan and processing done on it here: https://drive.google.com/file/d/1-ThagYTejkLouKWOXONSm41e22x30-1A/view?usp=sharing. I used the following command for registration: sudo sct_register_to_template -i mFFE_crop.nii -s mFFE_crop_seg.nii -l labels_vert.nii.gz -c t2 -param step=1,type=seg,algo=affine,metric=MeanSquares,gradStep=0.1,smooth=0,iter=50:step=2,type=seg,algo=affine,metric=MeanSquares,smooth=0,gradStep=0.1,iter=25:step=3,type=im,algo=syn,metric=MeanSquares,smooth=0,gradStep=0.25,iter=25 -ref subject -qc ~/Documents/Patient_Data_Folder/88/DTI_Data/niftis.
Thanks,
Ashwin
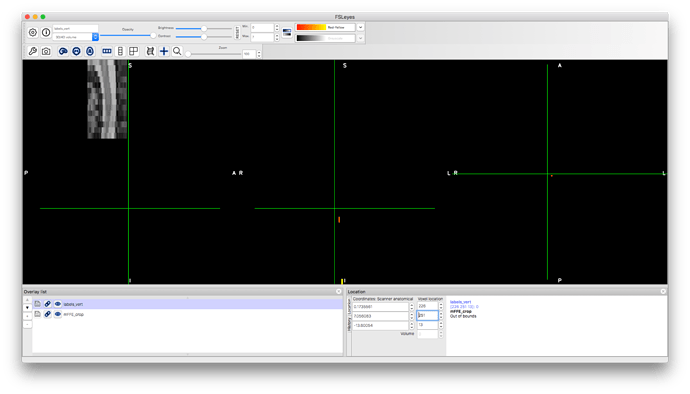
your labels are placed way below the FOV, see below (yellow and orange):
Dr. Cohenadad,
I used mipav to label the discs so that I could see the axial slices and place the labels on the centerline but it apparently didn’t label in the appropriate space. If I use sct_label_utils with -create-viewer, then it labels and registers in the appropriate FOV. Is there a way to see the axial slices using sct_labels_utils -create-viewer instead of the sagittal axis? Also, I would need to follow sct_labels_utils with sct_label_vertebrae to center the labeling, correct?
Thanks,
Ashwin
Hi,
you don’t need precision in the axial plane because this labeling is only used to fetch the z value corresponding to the level (the center of the cord in the axial plane is taken care of by the cord segmentation).
Thus, it is typically recommended to perform vertebral/disc labeling using an image where we can clearly identify the discs, which is not the case in this mFFE image. If you send me an anatomical image (e.g., T1w, T2w, etc.) corresponding to this subject, I can write a short pipeline for you
I don’t have access to the anatomical image. I instead found a workaround by modifying saggital.py. Thank you for your help.