Hi Ashwin,
The segmentation looks good, but the labeling seems off. First of all, the file is called labels_vert.nii.gz but in the syntax you used -ldisc, which is referring to disc labels (not vertebrae labels, as the file name seems to suggest).
Also, the top and bottom labels are respectively 2 (C2) and 8 (T1), and the volume has 10 slices of 5mm each, suggesting that C2 and T1 are separated by about 50mm which seems very small (usually in adult this distance would be about 10cm).
Lastly, and most importantly, the syntax uses -ref template (default behaviour), however the input file is highly anisotropic resolution (0.3x0.3x5mm), and as mentioned at page 59 of the SCT course, when inputting those kind of voxel resolution you absolutely need to avoid registering your subject to the template (-ref template), which involves a straightening procedure, and instead register the template to your subject (-ref subject). And when using -ref subject, only two labels can be input (as opposed to 7 in labels_vert.nii.gz).
Also, when using algo=syn,metric=MeanSquares, make sure the source and destination images have the same contrast and signal intensity (which is not the case here, between the mFFE and the PAM50_t2 images).
I tweaked a bit the registration parameters to accommodate for the large deformation of the spinal cord and not put too many iterations on the step that uses type=im to minimize spurious deformations driven by the pathological hyperintensities in the cord parenchyma (see syntax below).
Lastly, I would recommend using PAM50_t2s in place of PAM50_t2 for the mFEE scan.
Here is a suggested list of commands:
# Create labels at C2 and T1 (please check these labels, their distance seem suspiciously small to me)
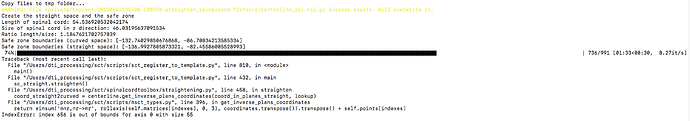
sct_label_utils -i mFFE_crop.nii -create-seg 0,8:10,2 -o mFFE_crop_labels_disc.nii.gz
# Register to template
sct_register_to_template -i mFFE_crop.nii -s mFFE_crop_seg.nii.gz -ldisc mFFE_crop_labels_disc.nii.gz -c t2s -ref subject -param step=1,type=seg,algo=centermass:step=2,type=seg,algo=bsplinesyn,gradStep=1,iter=25,slicewise=1:step=3,type=im,algo=syn,metric=CC,slicewise=1,iter=5 -qc qc-reg
Here is the QC report: qc-reg.zip (366.3 KB)
I hope it helps,
Julien