Hi,
I would like to know which warping fields (/tmp//warp) should I use to get a straight/LR symmetrical SC in PAM50 space (Step 1 to Step 5) without the non-rigid xy transformation (Step 6) and with only 1 interpolation?
Thank you,
Mounir M El Mendili
Hi Mounir,
If you keep the temporary folders on running sct_register_to_template (using flag -r 0), go to the temp folder, which is indicated in the STDOUT of your Terminal, example (see last line):
julien-macbook:~/sct_course_data/single_subject/data/t2 $ sct_register_to_template -i t2.nii.gz -s t2_seg.nii.gz -ldisc t2_labels_vert.nii.gz -c t2 -r 0
--
Spinal Cord Toolbox (git-master-3ce19094c99ea201fdc6757792e566ca3d7a9bb8)
sct_register_to_template -i t2.nii.gz -s t2_seg.nii.gz -ldisc t2_labels_vert.nii.gz -c t2 -r 0
--
Check template files...
OK: /Users/julien/code/spinalcordtoolbox/data/PAM50/template/PAM50_t2.nii.gz
OK: /Users/julien/code/spinalcordtoolbox/data/PAM50/template/PAM50_label_disc.nii.gz
OK: /Users/julien/code/spinalcordtoolbox/data/PAM50/template/PAM50_cord.nii.gz
Check parameters:
Data: t2.nii.gz
Landmarks: t2_labels_vert.nii.gz
Segmentation: t2_seg.nii.gz
Path template: /Users/julien/code/spinalcordtoolbox/data/PAM50
Remove temp files: 0
Check input labels...
Creating temporary folder (/var/folders/qd/2g7h7921023d_rzm472bf8jm0000gn/T/sct_2025-12-10_11-54-25_register-to-template_mt5dw3w0)
The warping field you’re interested in is called warp_curve2straightAffine.nii.gz.
See what it produces after running:
sct_apply_transfo -i data.nii -d template.nii -w warp_curve2straightAffine.nii.gz
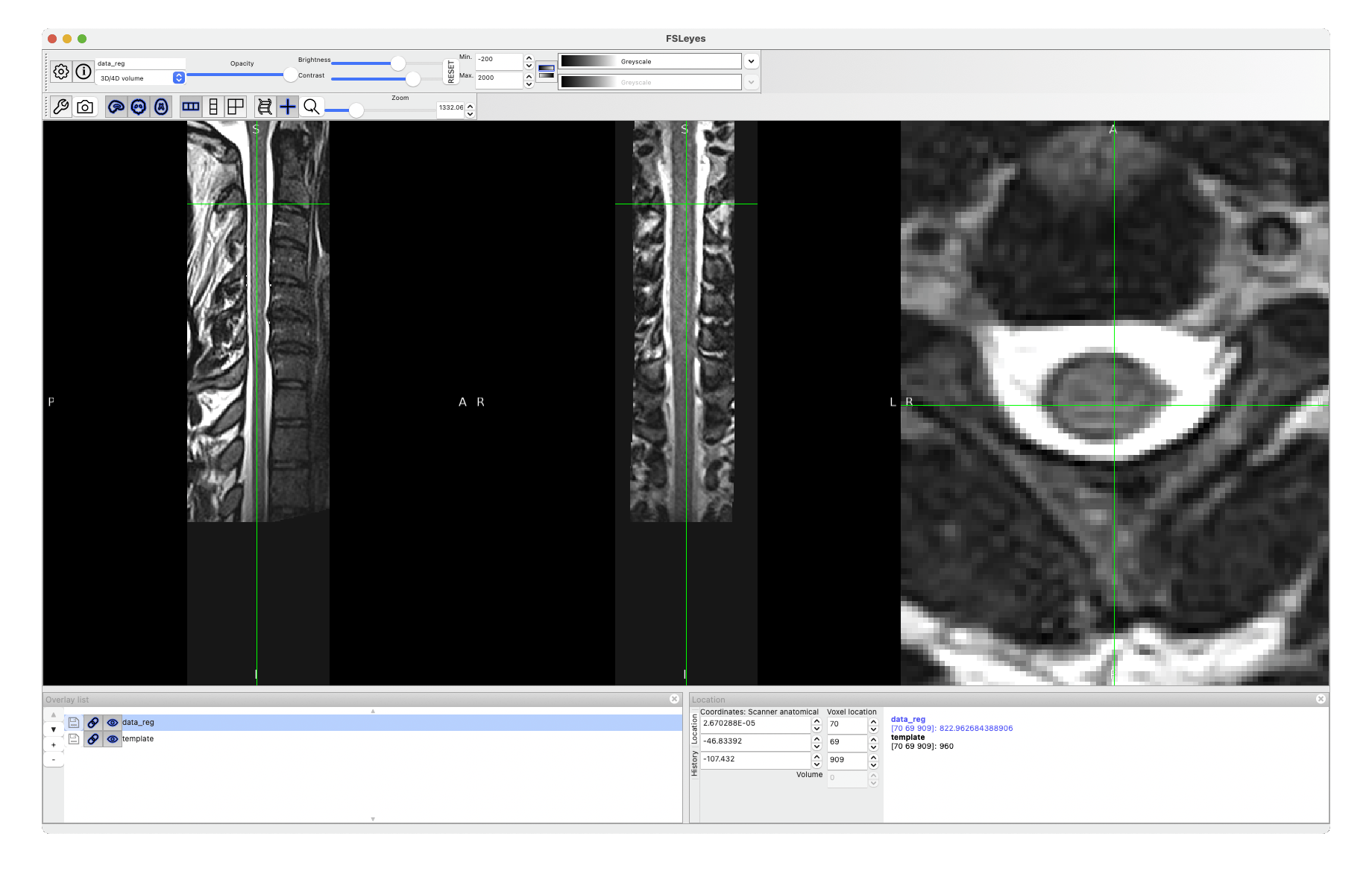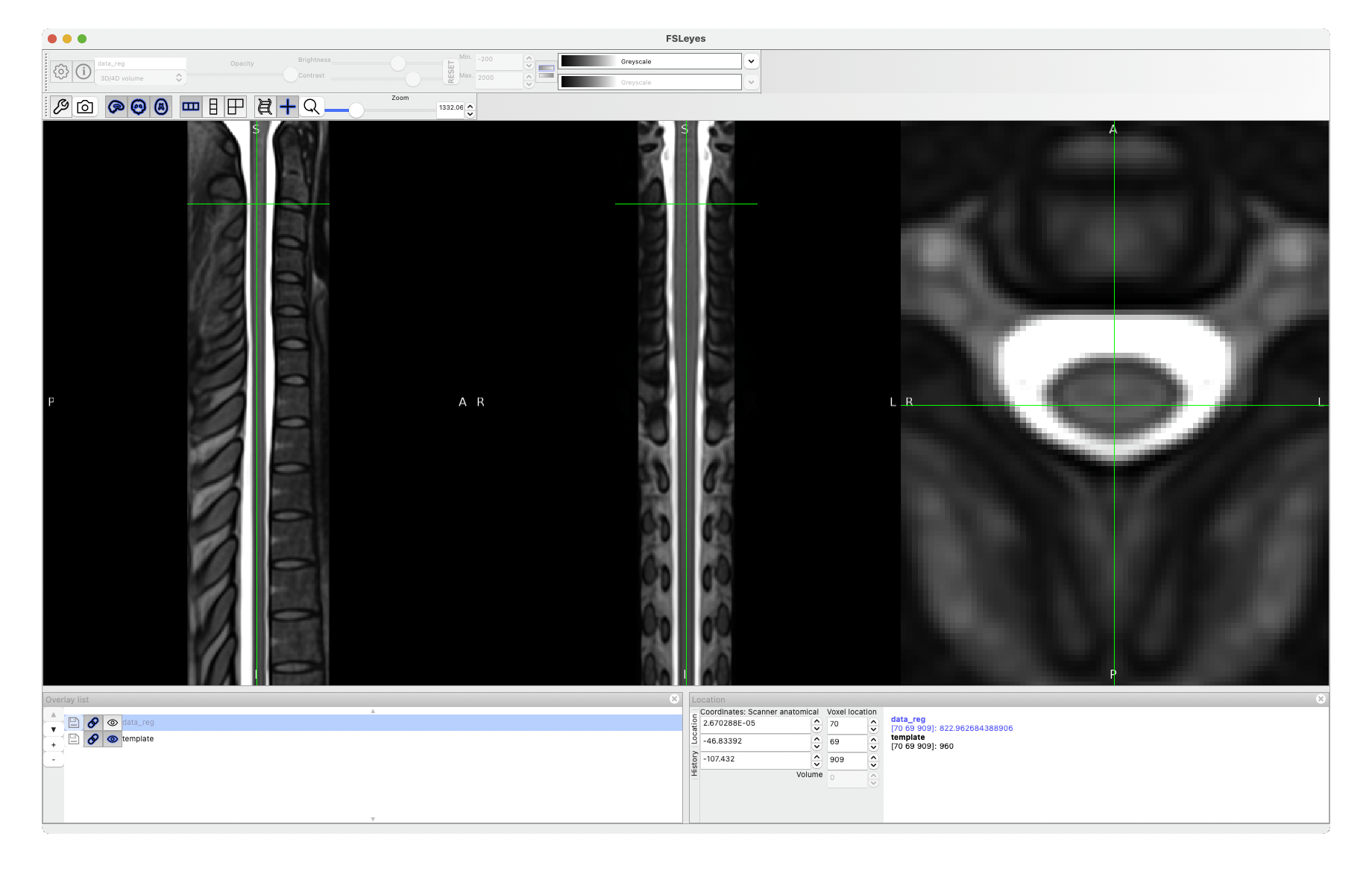
Cheers,
Julien
I’m trying to figure out which warp field should I use to transform the PAM50 template into the curve2straightAffine space. My current assumption is to use: warp_template_crop_sub2data_rpi_1mm_straightAffine_crop_sub.nii.gz
when I apply this warp, the overlap between warped PAM50 and curve2straightAffine_t2w is not that good. The terminal instructions seem straightforward, but I’m not sure if I’m interpreting them correctly?
Thank you very much!
Mounir
warp_template_crop_sub2data_rpi_1mm_straightAffine_crop_sub.nii.gz is a temporary warping field that uses a cropped version of the PAM50 template, which is done to accelerate processing. So no, you cannot apply that warping field on the PAM50 native template.
Maybe you can tell us exactly what you are trying to achieve?
I would prefer to analyze my cohort in an intermediate/common space rather than using the PAM50 space, because the following SyN registration would apply a very strong deformation on my images.
→ I want to bring the PAM50 template to that common space so I can use WM/GM probabilistic segmentations and WM atlases.
Is it possible to have the cropped version of the PAM50 template/the cropping window coordinates?
Thanks for your help!
Mounir
If you just want to skip the last non-linear deformations, you can simply customize the flag -param to specify only one step without iteration (equivalent of just running step=0), example:
sct_register_to_template -i t2.nii.gz -s t2_seg.nii.gz -l t2_labels_vert.nii.gz -c t2 -param step=1,type=seg,algo=syn,iter=0