Dear SCT team,
I tried to register my sample to template and an error occurs:
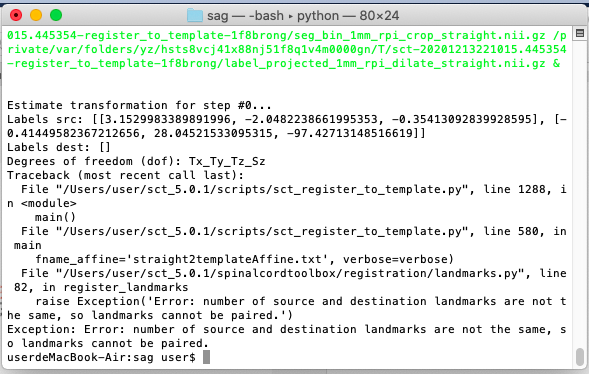
Exception: Error: number of source and destination landmarks are not the same, so landmarks cannot be paired.
What might be the problem and how should I correct it?
Please check the attached files and I am grateful for your help.
t2.nii.gz (2.3 MB)
t2_seg.nii.gz (18.5 KB)
Hi @ctchou,
Could you please copy/paste the syntax you typed, including the whole output, instead of only a screenshot (so that we can see what version of SCT you have, what input you used, etc.).
Also, you will need to upload the label (your input of -l or -ldisc) so I can try to reproduce your error.
Cheers,
Julien
Dear Julien,
Thank you for your kind response. Sorry for my unfamiliar with Linux and python.
I use the 5.0.1 edition
I used the default syntax:
sct_label_vertebrae -i t2.nii.gz -s t2_seg.nii.gz -c t2 -qc ~/qc_singleSubj
The result of labeling seemed a little bit deviation (please see the file)
t2_seg_labeled.nii.gz (111.8 KB)
but I continued to do the following step:
sct_label_utils -i t2_seg_labeled.nii.gz -vert-body 3,9 -o t2_labels_vert.nii.gz
t2_labels_vert.nii.gz (102.9 KB)
And I did the next procedure
sct_register_to_template -i t2.nii.gz -s t2_seg.nii.gz -l t2_labels_vert.nii.gz -c t2 -qc ~/qc_singleSubj
And I had the above error message.
I also tried to label manually:
sct_label_utils -i t2.nii.gz -create-viewer 3,9 -o t2_labels_disc_manual.nii.gz
t2_labels_disc_manual.nii.gz (25.9 KB)
and did the same procedure:
sct_register_to_template -i t2.nii.gz -s t2_seg.nii.gz -l t2_labels_disc_manual.nii.gz -c t2 -qc ~/qc_singleSubj
And I also got the error message.
What is my mistake and should I offer more information?
Thank you for your help again.
Hi,
Good news, I am able to reproduce your error. For full details, the error is caused by the manual labels not being recognized as integer:
julien-macbook:~/temp/20201216/new $ sct_register_to_template -i t2.nii.gz -s t2_seg.nii.gz -ldisc t2_labels_disc_manual.nii.gz -c t2 -qc qc
--
Spinal Cord Toolbox (git-lr/missing_disc_label_vertebrae-5bb113c5a7f17b10eadc9a1ebb01f50b94514f53)
sct_register_to_template -i t2.nii.gz -s t2_seg.nii.gz -ldisc t2_labels_disc_manual.nii.gz -c t2 -qc qc
--
Check template files...
OK: /Users/julien/code/sct/data/PAM50/template/PAM50_t2.nii.gz
OK: /Users/julien/code/sct/data/PAM50/template/PAM50_levels.nii.gz
OK: /Users/julien/code/sct/data/PAM50/template/PAM50_cord.nii.gz
Check parameters:
Data: t2.nii.gz
Landmarks: t2_labels_disc_manual.nii.gz
Segmentation: t2_seg.nii.gz
Path template: /Users/julien/code/sct/data/PAM50
Remove temp files: 1
Check input labels...
ERROR: Label should be integer.
ERROR: Label should be integer.
I’m investigating the issue and will come back to you shortly.
The issue was identified and fixed in https://github.com/neuropoly/spinalcordtoolbox/pull/3104.
Once the pull request is merged, we will create a patch (v5.0.2), so you will be able to download the new version and use it for your analyses.
Dear Julien,
Thank you for your kind assistance. I can keep my analysis again.
Best Regards