Dear SCT Team,
I am experiencing a challenge while using the sct_run_batch command in SCT version 6.0, particularly concerning the identification and processing of manually corrected segmentation file.
In cases where the script bypasses the step of locating the manually corrected segmentation file (due to the file being saved in an incorrect directory), the code executes smoothly. However, when I correctly save the manually corrected file in the designated directory, and it is successfully identified by the script, I encounter an IndexError during the execution of the sct_register_to_template command. This suggests a potential mismatch in the expected data dimensions or indices.
Attached, you will find the log file from my most recent run, along with the dimensions of the involved image files. I hope this information helps in diagnosing the issue. I would greatly appreciate any guidance or suggestions you might have to resolve this problem.
Thank you,
Fauziyya
err.preprocess_spinal_cord_new_sub-CSM070_ses-spinalcord.log (23.8 KB)
Hi @Mfauziyya,
My apologies, but it is difficult for me to diagnose your issue without being able to test the command using your data.
Are you able to share the following files:
Data: sub-CSM070_ses-spinalcord_T2w.nii.gz
Landmarks: sub-CSM070_ses-spinalcord_T2w_seg_labeled_discs_1to9.nii.gz
Segmentation: sub-CSM070_ses-spinalcord_T2w_seg.nii.gz
Kind regards,
Joshua
Hi Joshua,
Upon further testing, I discovered that when I use a segmentation file generated by sct_run_batch as a stand-in for a manually corrected file, the registration step succeeds without errors. This finding suggests that the root of the problem might be related to the segmentation and manual correction process, particularly since they were initially performed with SCT version 5.2.
I am sharing all the relevant segmentation files with you. These files are labeled according to how they were generated. My primary concern is to determine whether there are any significant differences between these files, especially considering they were produced using different versions of SCTAny guidance or suggestions on how to rectify this would be immensely helpful.
I have the segmentation files in question, labeled and the code including the log.
sub-CSM070_ses-spinalcord_T2w_seg_batchrun.nii.gz (36.6 KB)
sub-CSM070_ses-spinalcord_T2w_seg_labeled_discs_1to9.nii.gz (195.8 KB)
sub-CSM070_ses-spinalcord_T2w_seg5.2.nii.gz (32.4 KB)
sub-CSM070_ses-spinalcord_T2w_seg6.0.nii.gz (35.0 KB)
1 Like
err.preprocess_spinal_cord_new_sub-CSM070_ses-spinalcord.log (125.0 KB)
preprocess_spinal_cord_new.sh (16.0 KB)
unfortunately, the sub-CSM070_ses-spinalcord_T2w.nii.gz is too large to attach here. I wink share a link shortly
Thank you so much for sharing! I will begin investigating immediately. 
Please can you share your email so I can send you the file?
Of course! Please send the file to joshua.newton@polymtl.ca. 
Please can you double check the email address, from my end it says its not valid.
Thanks
Very strange! The email address should be correct?
I share the data using your gmail.
1 Like
Thank you! I have received the data. 
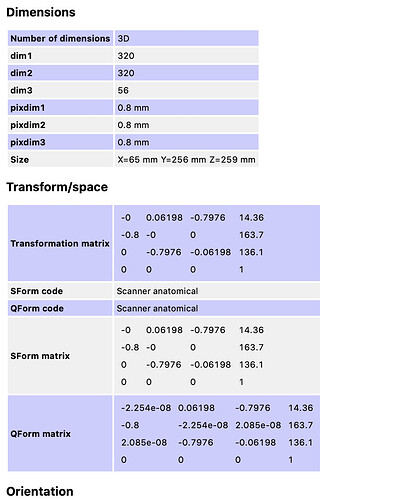
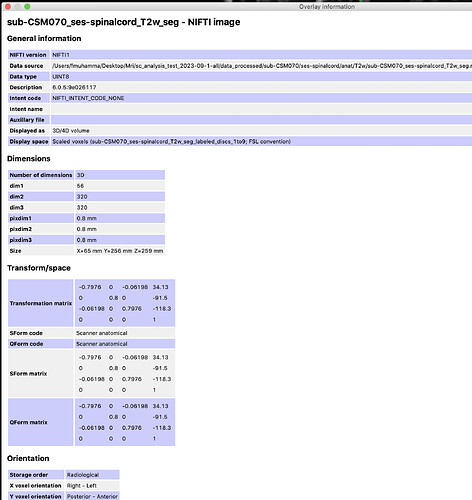
The first thing I notice is that the anat image, _seg_labeled_discs, and _seg_batchrun are all in ASR orientation, but _seg5.2 and _seg6.0 are both in RPI orientation.
So, I tried reorienting the segmentation using this command:
sct_image -setorient ASR -i sub-CSM070_ses-spinalcord_T2w_seg6.0.nii.gz -o sub-CSM070_ses-spinalcord_T2w_seg6.0_ASR.nii.gz
And, registration no longer fails when using the 6.0_ASR image. 
This is a strange bug, though. I would expect sct_register_to_template to internally reorient all of the input images to the same orientation before any processing. So, I don’t quite understand why reorienting the segmentation to ASR fixes the issue. I will need to do some further investigation in order to improve things on SCT’s end. I’ll follow up once I’ve identified the root cause here.
But, hopefully the command above solves things for you in the short term.
Kind regards,
Joshua
Hi Joshua,
Thank you very much for your assistance. I will reorient the manually corrected files for other subjects. I will reach out again oof the problem persist or if I encounter another error.
Kind regards,
Fauziyya
1 Like
Hi @Mfauziyya,
I have identified the root cause of the error here: Registration fails due to wrong cropping when seg/anat images have different orientations · Issue #4299 · spinalcordtoolbox/spinalcordtoolbox · GitHub
We will fix this issue in the next version of SCT so that orientation does not need to matter. But, for now, all you have to do is make sure that your labels and your segmentation are in the same orientation prior to registration.
Kind regards,
Joshua
Hi Joshua,
Excellent session today. I have emailed you my code.
Hi @Mfauziyya,
I have received your email! I will take a look after today’s session has completed. 
Kind regards,
Joshua
Hi Joshua,
Just checking in, I have updated my code based on the recommendation from today’s training. Please see attached code.
Please let me know what you think.
Fauziyya
My apologies, but I don’t see any attachments on your reply.
(The only code I have at the moment is the preprocess_spinal_cord_new.sh script that was sent via email on Monday, November 20, 2023 at 1:11 PM.)
Is there a newer script I should be looking at?
Kind regards,
Joshua
preprocess_spinal_cord_new.sh (20.9 KB)
Hi Joshua,
I have attached the code here. I emailed you this code yesterday as well.
Fauziyya
1 Like