Hi all,
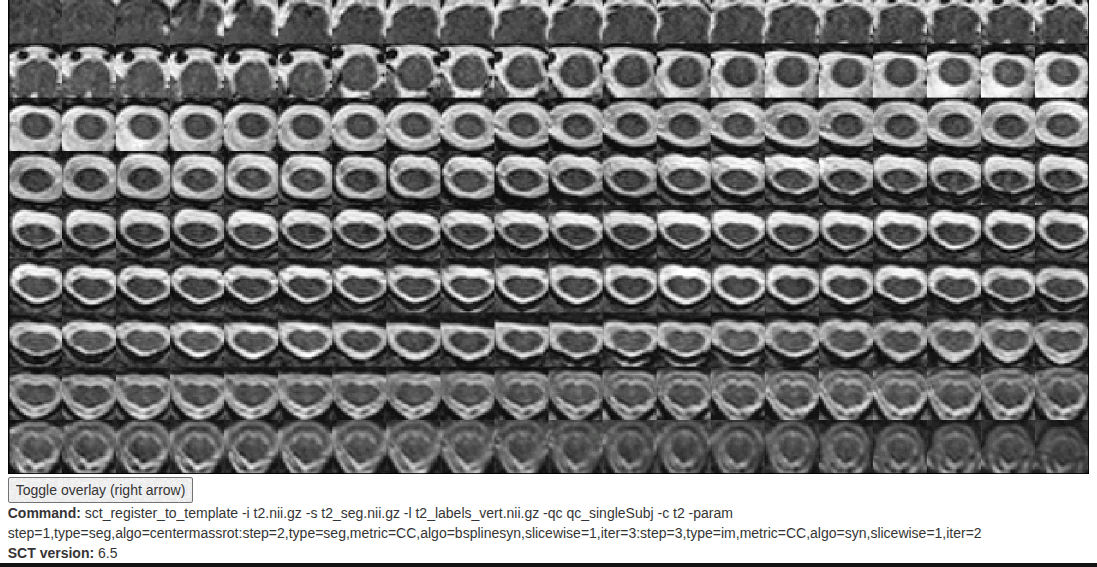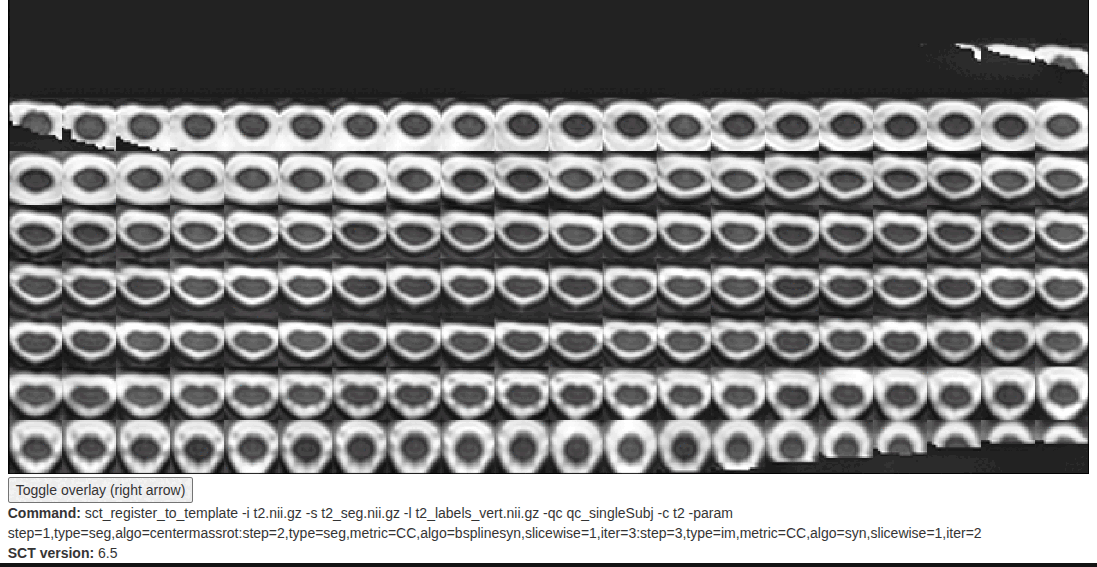
I am looking for suggestions to improve my SCT sct_register_to_template results. I have attached the QC folder for reference. Despite trying several options, the registration is still somewhat suboptimal. One specific issue I noticed is that the registered template (template2anat.nii.gz) often appears larger than my subject T2 (t2.nii.gz) spinal cord.
Here’s what I have tried so far:
Using vertebral labels 3–7
- Command:
sct_register_to_template -i t2.nii.gz -s t2_seg.nii.gz -l t2_labels_vert.nii.gz \
-qc qc_singleSubj -c t2 \
-param step=1,type=seg,algo=centermassrot:step=2,type=seg,metric=CC,algo=bsplinesyn,slicewise=1,iter=3:step=3,type=im,metric=CC,algo=syn,gradStep=0.2,smoothWarp=1.0,slicewise=1,iter=2
- Command (default parameters):
sct_register_to_template -i t2.nii.gz -s t2_seg.nii.gz -l t2_labels_vert.nii.gz -c t2 -qc qc_singleSubj
- Command (slightly modified parameters):
sct_register_to_template -i t2.nii.gz -s t2_seg.nii.gz -l t2_labels_vert.nii.gz \
-qc qc_singleSubj -c t2 \
-param step=1,type=seg,algo=centermassrot:step=2,type=seg,metric=CC,algo=bsplinesyn,slicewise=1,iter=3:step=3,type=im,metric=CC,algo=syn,slicewise=1,iter=2
- Command (more iterations and adjusted gradStep / smoothWarp):
sct_register_to_template -i t2.nii.gz -s t2_seg.nii.gz -l t2_labels_vert.nii.gz \
-qc qc_singleSubj -c t2 \
-param step=1,type=seg,algo=centermassrot:step=2,type=seg,metric=CC,algo=bsplinesyn,slicewise=1,iter=5,gradStep=0.25,smoothWarp=1.0:step=3,type=im,metric=CC,algo=syn,slicewise=1,iter=3,gradStep=0.15,smoothWarp=1.2
Using all disc labels
- Command (default parameters):
sct_register_to_template -i t2.nii.gz -s t2_seg.nii.gz -ldisc t2_seg_labeled_discs.nii.gz -c t2 -qc qc_singleSubj
- Command (complex parameters):
sct_register_to_template -i t2.nii.gz -s t2_seg.nii.gz -ldisc t2_seg_labeled_discs.nii.gz \
-qc qc_singleSubj -c t2 \
-param step=1,type=seg,algo=centermassrot:step=2,type=seg,metric=CC,algo=bsplinesyn,slicewise=1,iter=5,gradStep=0.25,smoothWarp=1.0:step=3,type=im,metric=CC,algo=syn,slicewise=1,iter=3,gradStep=0.15,smoothWarp=1.2
Additional information
- I used SCT v6.5 mainly because the segmentation tends to give slightly larger cord masks than v7.1, but I have tried both versions.
- Voxel size: 0.8 × 0.8 × 0.8 mm
- Cord coverage: C1–T1
Questions
- How good can
sct_register_to_templaterealistically get for high-resolution isotropic T2 data? - Are there any tips or best practices to assess registration quality quantitatively and visually?
- Do you have suggestions for parameter adjustments or preprocessing steps that could improve template alignment and prevent AP cord expansion?
Thanks in advance for any guidance!
Best,
Jia
qc_singleSubj.zip (4.8 MB)