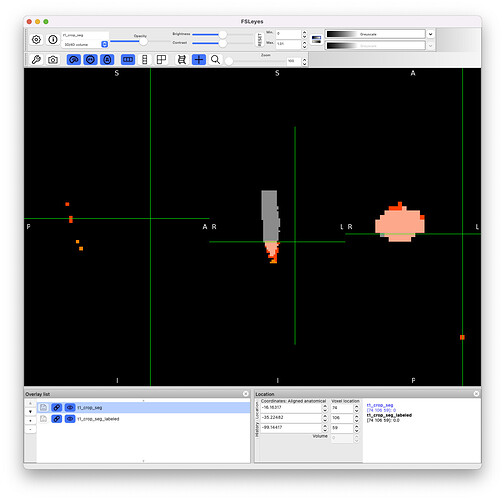
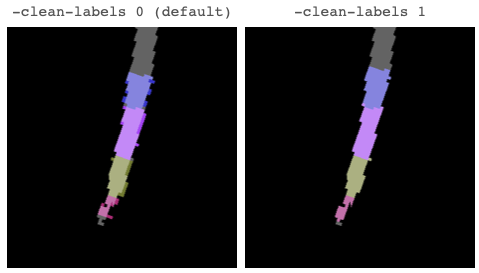
1. A description of the problem: When reviewing the csa.csv output using the whole/cropped T1 as input, we needed some spurious slices being included in the calculations. For example, C1:C2 average used slices 33:60;62:63;65;75:76, rather than 33:60. Upon visual inspection, I noted: (a) spurious voxels in the label_seg away from the spinal cord (seg image); and (b) voxels in the label_seg image adjacent to the cord and also not seg image. I was expecting that the label_seg would be constrained to only include voxels in the seg mask image. This does not happen in all cases and doesn’t happen with test case on this forum using the whole brain.
2. Commands and terminal output.
sct_deepseg_sc -i t1_crop.nii -c t1 -brain 1 -centerline cnn;
sct_label_vertebrae -i t1_crop.nii -s t1_crop_seg.nii -c t1;
sct_process_segmentation -i t1_crop_seg.nii -vertfile t1_crop_seg_labeled.nii -vert 1:2 -o my_csa.csv -append 1;
3. System information.
Spinal Cord Toolbox (5.5)
sct_check_dependencies
SCT info:
- version: 5.5
- path: /Users/dmclaren/sct_5.5
OS: osx (Darwin-20.6.0-x86_64-i386-64bit)
CPU cores: Available: 12, Used by ITK functions: 12
RAM: Total: 16384MB, Used: 10034MB, Available: 5259MB
Check Python executable…[OK]
Using bundled python 3.7.11 (default, Jul 27 2021, 07:03:16)
[Clang 10.0.0 ] at /Users/dmclaren/sct_5.5/python/envs/venv_sct/bin/python
Check if data are installed…[OK]
Check if colored is installed…[OK] (1.4.3)
Check if dipy is installed…[OK] (1.4.1)
Check if h5py is installed…[OK] (2.10.0)
Check if Keras (2.3.1) is installed…[OK] (2.3.1)
Check if ivadomed is installed…[OK] (2.9.2)
Check if matplotlib is installed…[OK] (3.5.1)
Check if nibabel is installed…[OK] (3.2.1)
Check if numpy is installed…[OK] (1.18.5)
Check if onnxruntime is installed…[OK] (1.7.0)
Check if pandas is installed…[OK] (1.3.5)
Check if psutil is installed…[OK] (5.9.0)
Check if pyqt5 (5.11.3) is installed…[OK] (5.11.3)
Check if pytest is installed…[OK] (6.2.5)
Check if pytest-cov is installed…[OK] (3.0.0)
Check if raven is installed…[OK]
Check if requests is installed…[OK] (2.27.1)
Check if requirements-parser is installed…[OK]
Check if scipy is installed…[OK] (1.7.3)
Check if scikit-image is installed…[OK] (0.19.1)
Check if scikit-learn is installed…[OK] (1.0.2)
Check if tensorflow is installed…[OK] (1.15.5)
Check if torch (1.5.0) is installed…[OK] (1.5.0)
Check if torchvision (0.6.0) is installed…[OK] (0.6.0)
Check if xlwt is installed…[OK] (1.3.0)
Check if tqdm is installed…[OK] (4.62.3)
Check if transforms3d is installed…[OK] (0.3.1)
Check if urllib3 is installed…[OK] (1.26.8)
Check if pytest_console_scripts is installed…[OK]
Check if wquantiles is installed…[OK] (0.4)
Check if spinalcordtoolbox is installed…[OK]
Check ANTs compatibility with OS …[OK]
Check PropSeg compatibility with OS …[OK]
Check if figure can be opened with matplotlib…[OK] (Using GUI backend: ‘MacOSX’)
Check if figure can be opened with PyQt…[OK]
Check FSLeyes version…Command not found
- File upload. (Please upload any data or scripts needed to reproduce your issue, if there are any.) →