Hi,
I would like to know how to compute SNR on a diffusion image in SCT. I have inspected sct_compute_snr but I could not understand what to put instead of -vol option.
best regards,
Rosella
Hi @Rosella_Tro,
Thank you for reaching out. So, let’s start with an example dMRI dataset:
# Download example data
sct_download_data -d sct_example_data
# Go to DWI data
cd sct_example_data/dmri
So, at this point there are different ways to compute SNR on a dMRI dataset. One way to do it, is to compute it on the b=0 images only, to remove bias from the diffusion-related signal attenuation, ie: you want to quantify thermal noise only.
To identify where the b=0 images are, you can run this:
sct_dmri_separate_b0_and_dwi -i dmri.nii.gz -bvec bvecs.txt
The output is:
[...]
Identify b=0 and DWI images...
WARNING: bvecs file is 3xn instead of nx3. Consider using sct_dmri_transpose_bvecs.
Transpose bvecs...
Number of b=0: 5 [0, 31, 32, 33, 34]
Number of DWI: 30 [1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30]
[...]
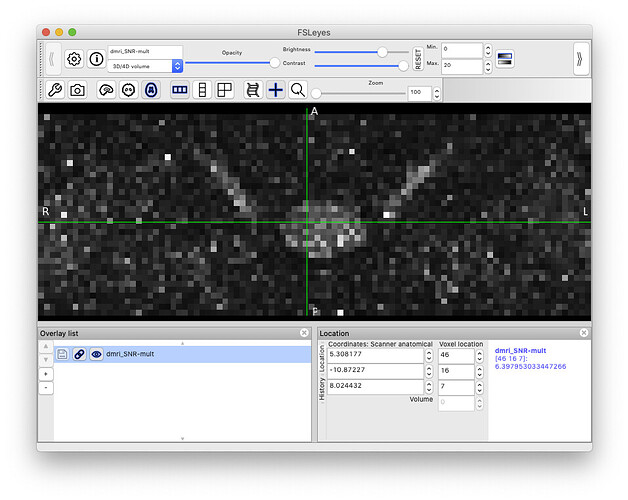
Now that you know where the b=0 images are, you can use them to compute SNR using the “mult” method, as described in [Dietrich et al. J Magn Reson Imaging 2007]. In brief, with this method, noise standard deviation (SD) is computed across time. So you need enough samples (ie: volumes) to compute a reliable SD. Here, we have 5, which is “borderline”.
sct_compute_snr -i dmri.nii.gz -method mult -vol 0,31:34
This outputs the file dmri_SNR-mult.nii.gz, which shows the voxel-wise SNR:
If you have a mask of the spinal cord, you could input it to extract SNR inside the cord. That’s what we are going to do here:
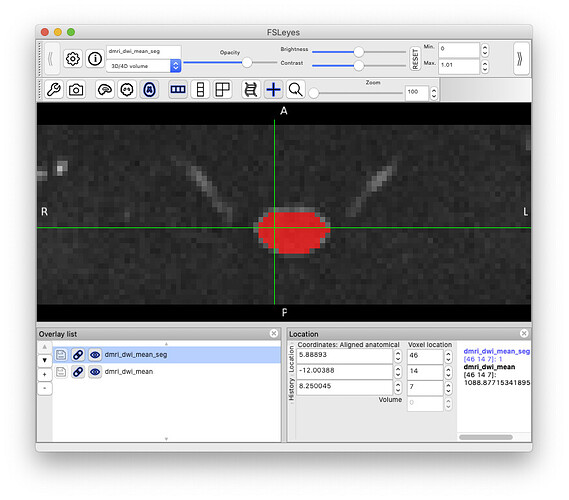
sct_deepseg_sc -i dmri_dwi_mean.nii.gz -c dwi -qc qc
Which gives us this spinal cord mask:
Which we can then use to extract average SNR in the spinal cord at each slice:
sct_extract_metric -i dmri_SNR-mult.nii.gz -f dmri_dwi_mean_seg.nii.gz -o snr_mult.csv
Output:
| Slice (I->S) | Size [vox] | WA() | STD() |
|---|---|---|---|
| 0 | 80 | 8.379061 | 5.376076701355050 |
| 1 | 81 | 7.6254206 | 2.9527455242505100 |
| 2 | 85 | 8.024328 | 7.179199467822250 |
| 3 | 91 | 8.088527 | 3.894383220097640 |
| 4 | 92 | 8.511543 | 4.6690126153706500 |
| 5 | 86 | 8.035383 | 3.9745487892725800 |
| 6 | 88 | 7.688621 | 3.20418832388819 |
| 7 | 87 | 7.3579865 | 3.135453706609150 |
| 8 | 82 | 7.112298 | 3.190911953956170 |
| 9 | 77 | 7.7654552 | 3.780912526101250 |
| 10 | 78 | 7.026523 | 3.4383660352603000 |
| 11 | 71 | 6.530175 | 2.9450315953682500 |
| 12 | 70 | 6.7164145 | 3.3545565803044500 |
| 13 | 75 | 7.267548 | 4.877266381484310 |
| 14 | 71 | 6.702518 | 2.739299068038260 |
Now, let’s look at another approach to compute SNR, which is based on the difference between two images and the computation of the noise SD within a spatial mask (as opposed to across time, as done with the “mult” method). With this method, you need to input a mask that specifies the region where noise SD is computed. In general, the mask is defined in a region of interest, and one assumption is that noise SD is invariant across voxels in the mask (this property is called homoscedasticity). Here, we will use the spinal cord mask:
sct_compute_snr -i dmri.nii.gz -m dmri_dwi_mean_seg.nii.gz -method diff -vol 31,32
Output:
SNR_diff = 5.891875075931879
When using the “diff” or “mult” methods, it is important that images are co-registered before computing SNR. Also, the interpolation used for motion correction should not be linear or sinc, because it will alter noise property. Interpolation should only be nearest-neighbour.
thank you as always for your clarity 
best,
Rosella