Dear Sunny,
Thank you for sharing your sample data and commands with me. I had to make some small corrections to the commands you provided in the .zip (i.e. t2-l.txt and dmri_2.txt), but I mainly just tried to re-run your commands and make observations:
T2 commands
Here are the commands I tried for the T2 data:
# segment the spinal cord
export QC_FOLDER="path/to/output/qc"
sct_deepseg -i t2.nii.gz -task seg_lumbar_sc_t2w -qc "$QC_FOLDER"
# create a labeled segmentation
sct_label_utils -i t2.nii.gz -create-viewer 19 -o label_T11T12.nii.gz -msg "Click at the posterior tip of T11/T12 inter-vertebral disc"
sct_label_vertebrae -i t2.nii.gz -s t2_seg.nii.gz -c t2 -initlabel label_T11T12.nii.gz -qc "$QC_FOLDER"
# extract vert body labels
sct_label_utils -i t2_seg_labeled.nii.gz -vert-body 19 -o t2_labels_vert.nii.gz
sct_qc -i t2.nii.gz -s t2_labels_vert.nii.gz -p sct_label_utils -qc "$QC_FOLDER"
# use the labels to register to template
sct_register_to_template -i t2.nii.gz -s t2_seg.nii.gz -l t2_labels_vert.nii.gz -qc "$QC_FOLDER" -ofolder advanced_param -c t2 -param step=1,type=seg,algo=rigid:step=2,type=seg,metric=CC,algo=bsplinesyn,slicewise=1,iter=3:step=3,type=im,metric=CC,algo=syn,slicewise=1,iter=2
sct_warp_template -d t2_lumbar.nii.gz -w advanced_param/warp_template2anat.nii.gz -a 0 -qc "$QC_FOLDER"
Some notes:
- To use the
QC_FOLDER variable, you need to use $ syntax ($QC_FOLDER or "$QC_FOLDER")
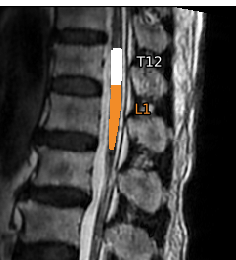
sct_label_vertebrae failed at first when using label 18 (label_T10T11.nii.gz). I had to switch to label 19 instead.- The
sct_label_vertebrae step excluded some of the top region of the cord segmentation (everything above the T12 vertebral level)
- The
sct_label_utils command included “-vert-body 17,19” though 17 didn’t exist.
- There were some commented-out commands identifying “label 60” (cauda equina). I have not tested your commands yet, but maybe “60” could be useful.
For sample subject 1, the T2 steps seem to go OK:
dMRI commands
Next, here are the commands I tried for the dMRI data:
# generate mask for moco
sct_dmri_separate_b0_and_dwi -i dmri.nii.gz -bvec bvecs.txt
sct_deepseg_sc -i dmri_dwi_mean.nii.gz -c dwi -qc "$QC_FOLDER"
sct_create_mask -i dmri_dwi_mean.nii.gz -p centerline,dmri_dwi_mean_seg.nii.gz -size 35mm
# apply motion correction
sct_dmri_moco -i dmri.nii.gz -m mask_dmri_dwi_mean.nii.gz -bvec bvecs.txt
# create "good" spinal cord mask on moco'd image
sct_deepseg -task seg_sc_contrast_agnostic -i dmri_moco_dwi_mean.nii.gz -qc "$QC_FOLDER"
# perform registration
sct_register_multimodal -i "${SCT_DIR}/data/PAM50/template/PAM50_t1.nii.gz" \
-iseg "${SCT_DIR}/data/PAM50/template/PAM50_cord.nii.gz" \
-d dmri_moco_dwi_mean.nii.gz \
-dseg dmri_moco_dwi_mean_seg.nii.gz \
-initwarp ../t2/advanced_param/warp_template2anat.nii.gz \
-initwarpinv ../t2/advanced_param/warp_anat2template.nii.gz \
-owarp warp_template2dmri.nii.gz \
-owarpinv warp_dmri2template.nii.gz \
-param step=1,type=seg,algo=centermass:step=2,type=seg,algo=bsplinesyn,slicewise=1,iter=3 \
-qc ~/qc_singleSubj
Some notes:
- The motion correction and “
sct_deepseg” step seemed to go (mostly) OK:
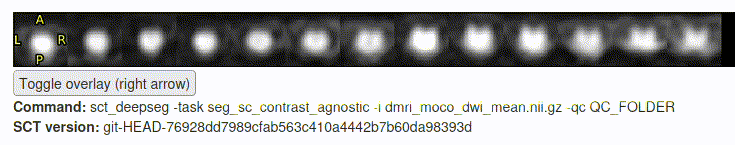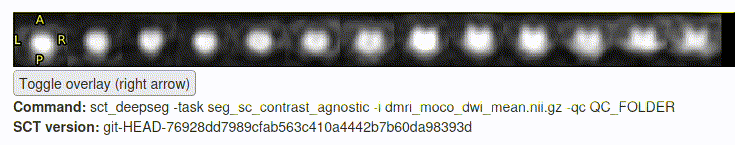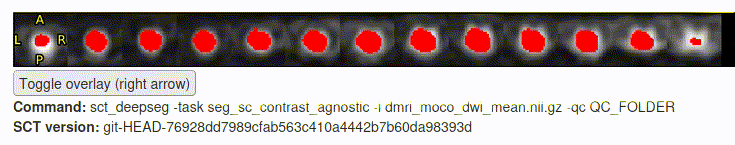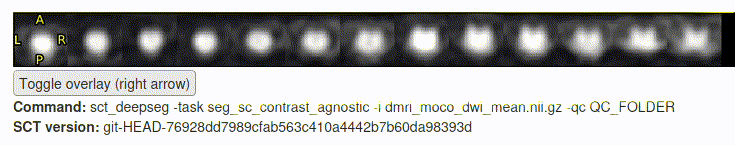
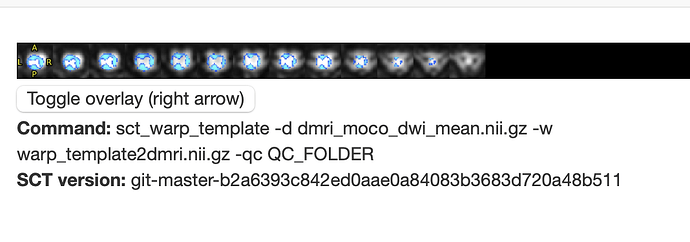
- However, the initial files
t2.nii.gz and dmri.nii.gz do not appear to be very well aligned:
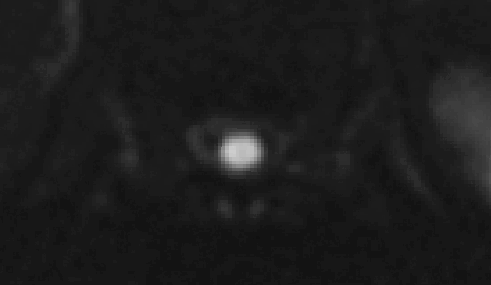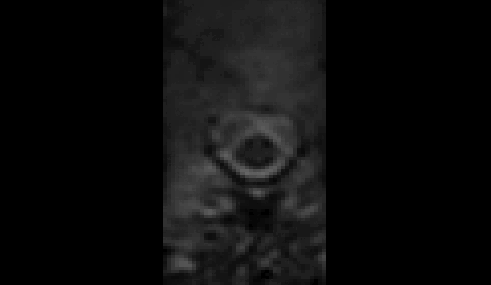
- So, when the T2 “
-initwarp” file gets applied to the dMRI image at the start of the registration, the dMRI segmentation is also mis-aligned:
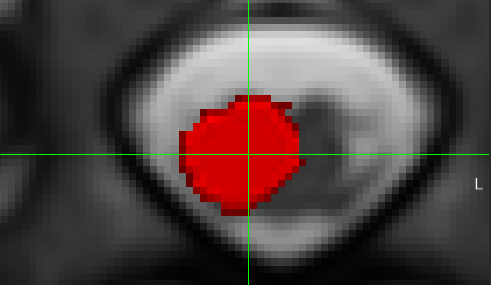
- For comparison, here is the sample data that SCT uses in its tutorials (
sct_tutorial_data):

Based on the above notes, I think the alignment between the T2 and dMRI data may need to be double-checked first, prior to running sct_register_multimodal between the dMRI and the template.
I will also ask my colleagues for further input. Maybe there is set of registration parameters that will work OK without an initial alignment, so I will ask them.
Thank you kindly for your patience and understanding,
Joshua