Hi
I recovered your post with the gdrive link (it was flagged as spam by the system).
I will download your data and have a look when I find some time, hopefully today
It would be great,
thank you,
Rosella
@Rosella_Tro the drive link is not there anymore. Could you please put it back? I removed the filter that spams posts with drive links in there.
sorry about the inconvenience
sorry for the delay in my answer but I have data in my office lab. I have tried to attach the google drive link but the post gets flagged by the community again. do you manage to recover that link from the spam?
thank you,
Rosella
Sorry it doesn’t seem to work— can you just email me the link?
@Rosella_Tro I did have a look at your data and came up with a dedicated workflow for this dataset, where part of the image should be ignored from the analysis. I’ve created a video tutorial which you can access here.
Below is the series of script that I’ve used for your data. As you can see, the sct_propseg line was replaced with sct_deepseg_sc, and includes manual initialization:
sct_dmri_separate_b0_and_dwi -i kurtosis.nii -bvec kurtosis_crop_moco.bvec
sct_deepseg_sc -i kurtosis_dwi_mean.nii -c dwi -centerline viewer
sct_create_mask -i kurtosis_dwi_mean.nii -p centerline,kurtosis_dwi_mean_seg.nii -size 30mm -o mask_kurtosis_dwi_mean.nii
sct_crop_image -i kurtosis.nii -m mask_kurtosis_dwi_mean.nii -o kurtosis_crop.nii
sct_dmri_moco -i kurtosis_crop.nii -bvec kurtosis_crop_moco.bvec -bval kurtosis_crop_moco.bval -x spline
sct_deepseg_sc -i kurtosis_crop_moco_dwi_mean.nii -c dwi
Hope that helps.
Best,
Julien
I expected I should use a manual initialization and I will try to fix the viewer function on my computer as probably some library is not installed. Anyway, thank you very much for the help. If you wish, I will update you with my results .
Thank you,
Rosella
When runnign the script you provide me, the following error raises:
Spinal Cord Toolbox (dev)
Config deepseg_sc:
_ Centerline algorithm: viewer_
_ Brain in image: False_
_ Kernel dimension: 2d_
_ Contrast: dwi_
_ Threshold: 0.01_
Create temporary folder (/tmp/sct-20191113102740.638922-xrk1ttx)…_
Reorient the image to RPI, if necessary…
Finding the spinal cord centerline…
Traceback (most recent call last):
_ File “/home/rosella/sct_dev/scripts/sct_deepseg_sc.py”, line 201, in _
_ main()_
_ File “/home/rosella/sct_dev/scripts/sct_deepseg_sc.py”, line 185, in main_
_ threshold_seg=threshold, remove_temp_files=remove_temp_files, verbose=verbose)_
_ File “/home/rosella/sct_dev/spinalcordtoolbox/deepseg_sc/core.py”, line 477, in deep_segmentation_spinalcord_
_ centerline_fname=file_ctr)_
_ File “/home/rosella/sct_dev/spinalcordtoolbox/deepseg_sc/core.py”, line 106, in find_centerline_
_ im_labels = call_viewer_centerline(im)
_ File “/home/rosella/sct_dev/spinalcordtoolbox/centerline/core.py”, line 268, in call_viewer_centerline
_ from spinalcordtoolbox.gui.base import AnatomicalParams_
_ File “/home/rosella/sct_dev/spinalcordtoolbox/gui/base.py”, line 18, in _
_ from PyQt5 import QtCore, QtGui, QtWidgets_
ImportError: dlopen: cannot load any more object with static TLS
–
I am currently searching how to solve it.
Thank you,
Rosella
hum… this might be a memory issue (might be related to this).
could you please run the following commands and copy/paste the output:
sct_check_dependencies
sct_testing
sct_check_dependencies
–
Spinal Cord Toolbox (4.1.0)
SCT info:
- version: 4.1.0
- path: /home/rosella/sct_4.1.0
OS: linux (Linux-3.16.0-4-amd64-x86_64-with-debian-8.11)
CPU cores: Available: 8, Used by SCT: 8
RAM: MemTotal: 32970652 kB
total used free shared buffers cached
Mem: 32197 6822 25374 77 359 3834
-/+ buffers/cache: 2628 29569
Swap: 57218 0 57218
Check Python executable…[OK]
Using bundled python 3.6.9 |Anaconda, Inc.| (default, Jul 30 2019, 19:07:31)
[GCC 7.3.0] at /home/rosella/sct_4.1.0/python/envs/venv_sct/bin/python
Check if data are installed…[OK]
Check if numpy is installed…[OK] (1.17.4)
Check if cryptography is installed…[OK] (2.8)
Check if dipy is installed…[OK] (1.0.0)
Check if futures is installed…[OK]
Check if h5py is installed…[OK] (2.10.0)
Check if Keras is installed…[OK] (2.3.1)
Check if matplotlib is installed…[OK] (3.1.1)
Check if nibabel is installed…[OK] (2.5.1)
Check if pandas is installed…[OK] (0.25.3)
Check if psutil is installed…[OK] (5.6.5)
Check if pyqt5 is installed…[OK]
Check if pytest is installed…[OK] (5.2.2)
Check if pytest-cov is installed…[OK] (2.8.1)
Check if raven is installed…[OK]
Check if requests is installed…[OK] (2.22.0)
Check if scipy is installed…[OK] (1.3.2)
Check if scikit-image is installed…[OK] (0.16.2)
Check if scikit-learn is installed…[OK] (0.21.3)
Check if tensorflow is installed…[OK] (2.0.0)
Check if xlrd is installed…[OK] (1.2.0)
Check if xlutils is installed…[OK]
Check if xlwt is installed…[OK] (1.3.0)
Check if tqdm is installed…[OK] (4.38.0)
Check if transforms3d is installed…[OK] (0.3.1)
Check if urllib3[secure] is installed…[OK] (1.25.7)
Check if spinalcordtoolbox is installed…[OK]
Check ANTs compatibility with OS …[OK]
Check PropSeg compatibility with OS …[OK]
Check if figure can be opened…[OK]
rosella@vivi:~$ sct_testing
–
Spinal Cord Toolbox (4.1.0)
Will run through the following tests:
- sequentially: sct_deepseg_gm sct_deepseg_lesion sct_deepseg_sc
- in parallel with 8 jobs: sct_analyze_lesion sct_analyze_texture sct_apply_transfo sct_convert sct_compute_ernst_angle sct_compute_hausdorff_distance sct_compute_mtr sct_compute_mscc sct_compute_snr sct_concat_transfo sct_create_mask sct_crop_image sct_dice_coefficient sct_detect_pmj sct_dmri_compute_dti sct_dmri_concat_b0_and_dwi sct_dmri_concat_bvals sct_dmri_concat_bvecs sct_dmri_create_noisemask sct_dmri_compute_bvalue sct_dmri_moco sct_dmri_separate_b0_and_dwi sct_dmri_transpose_bvecs sct_extract_metric sct_flatten_sagittal sct_fmri_compute_tsnr sct_fmri_moco sct_get_centerline sct_image sct_label_utils sct_label_vertebrae sct_maths sct_merge_images sct_process_segmentation sct_propseg sct_qc sct_register_multimodal sct_register_to_template sct_resample sct_smooth_spinalcord sct_straighten_spinalcord sct_warp_template
Checking sct_deepseg_gm…[OK]
Checking sct_deepseg_lesion…[OK]
Checking sct_deepseg_sc…[OK]
Checking sct_analyze_lesion…[OK]
Checking sct_analyze_texture…[OK]
Checking sct_apply_transfo…[OK]
Checking sct_convert…[OK]
Checking sct_compute_ernst_angle…[OK]
Checking sct_compute_hausdorff_distance…[OK]
Checking sct_compute_mtr…[OK]
Checking sct_compute_mscc…[OK]
Checking sct_compute_snr…[OK]
Checking sct_concat_transfo…[OK]
Checking sct_create_mask…[OK]
Checking sct_crop_image…[OK]
Checking sct_dice_coefficient…[OK]
Checking sct_detect_pmj…[OK]
Checking sct_dmri_compute_dti…[OK]
Checking sct_dmri_concat_b0_and_dwi…[OK]
Checking sct_dmri_concat_bvals…[OK]
Checking sct_dmri_concat_bvecs…[OK]
Checking sct_dmri_create_noisemask…[OK]
Checking sct_dmri_compute_bvalue…[OK]
Checking sct_dmri_moco…[OK]
Checking sct_dmri_separate_b0_and_dwi…[OK]
Checking sct_dmri_transpose_bvecs…[OK]
Checking sct_extract_metric…[OK]
Checking sct_flatten_sagittal…[OK]
Checking sct_fmri_compute_tsnr…[OK]
Checking sct_fmri_moco…[OK]
Checking sct_get_centerline…[OK]
Checking sct_image…[OK]
Checking sct_label_utils…[OK]
Checking sct_label_vertebrae…[OK]
Checking sct_maths…[OK]
Checking sct_merge_images…[OK]
Checking sct_process_segmentation…[OK]
Checking sct_propseg…[OK]
Checking sct_qc…[OK]
Checking sct_register_multimodal…[OK]
Checking sct_register_to_template…[OK]
Checking sct_resample…[OK]
Checking sct_smooth_spinalcord…[OK]
Checking sct_straighten_spinalcord…[OK]
Checking sct_warp_template…[OK]
status: [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0]
Finished! Elapsed time: 112s
Thanks, when you run the following command, do you see a window popping up?
sct_label_utils -i kurtosis_dwi_mean.nii -create-viewer 1
If not, could you copy/paste then entire terminal output (including the syntax)?
Are you running SCT remotely by connecting via ssh to a server?
rosella@vivi:~/1/Rm_Encefalo/DKI_b700_b2100_6b0$ sct_label_utils -i kurtosis_dwi_mean.nii -create-viewer 1
–
Spinal Cord Toolbox (4.1.0)
/home/rosella/sct_4.1.0/python/envs/venv_sct/bin/python: relocation error: /home/rosella/sct_4.1.0/python/envs/venv_sct/lib/python3.6/site-packages/PyQt5/Qt/plugins/platforms/…/…/lib/libQt5DBus.so.5: symbol dbus_message_get_allow_interactive_authorization, version LIBDBUS_1_3 not defined in file libdbus-1.so.3 with link time reference
I am using SCT on my PC without connection via ssh
Rosella
OK, I will need to do some investigations by trying to reproduce your OS setup. Sorry about the inconvenience.
Do not worry, is there some information I can provide in order to help?
thank you,
Rosella
Hi,
Waiting for resolving the problem with the viewer I found a temporary solution which consists in using deepseg with a centerline file provided by the user and created in fsl. This way I manage to segment the image, to crop it and compute subsequent DTI and DKI metrics. Segmentation is not optimal since it depends on the mask I created manually, and DTI and DKI maps have some white or black voxels which I will have to remove somehow, but this is probably due to noise.
The next issue is to provide a gray and white matter segmentation from b0 images. I have tried with deepseg_gm but results are poor. Is there a way to compute automatic segmentation from b0 images, even if the contrast between grey and white matter is poor?
thank you ,
Rosella
ps. Please find attached the script I used and a link with all output images.script_soggetti_spinale.sh (8.9 KB)
https://drive.google.com/drive/u/0/folders/1ZPG8B0Yo8ySpcG_L3ltPv9hg67HnEfxs
Hi,
I strongly advise against running sct_deepseg_gm on your b=0 DWI data. Even with the naked eye, we don’t see the GM. It would produce spurious results that you should not trust.
Instead, I suggest you to rely on the average GM mask (part of the PAM50 template), which can be warped after you run sct_register_to_template.
Thank you very much,
In the meantime I continue trying to solve the problem with the viewer.
Best regards,
Rosella
How can I resolve the problem of spurious (black and white) voxels inside my DKI or DTI maps?
Is this a problem of noise related to the quality of my images? is there anything I can do?
Here is a link to the maps:
https://drive.google.com/drive/u/0/folders/1Ow7zh9wxHv7Tqa74MQ7gEnuyNQ9OD16N
Thank you
Rosella
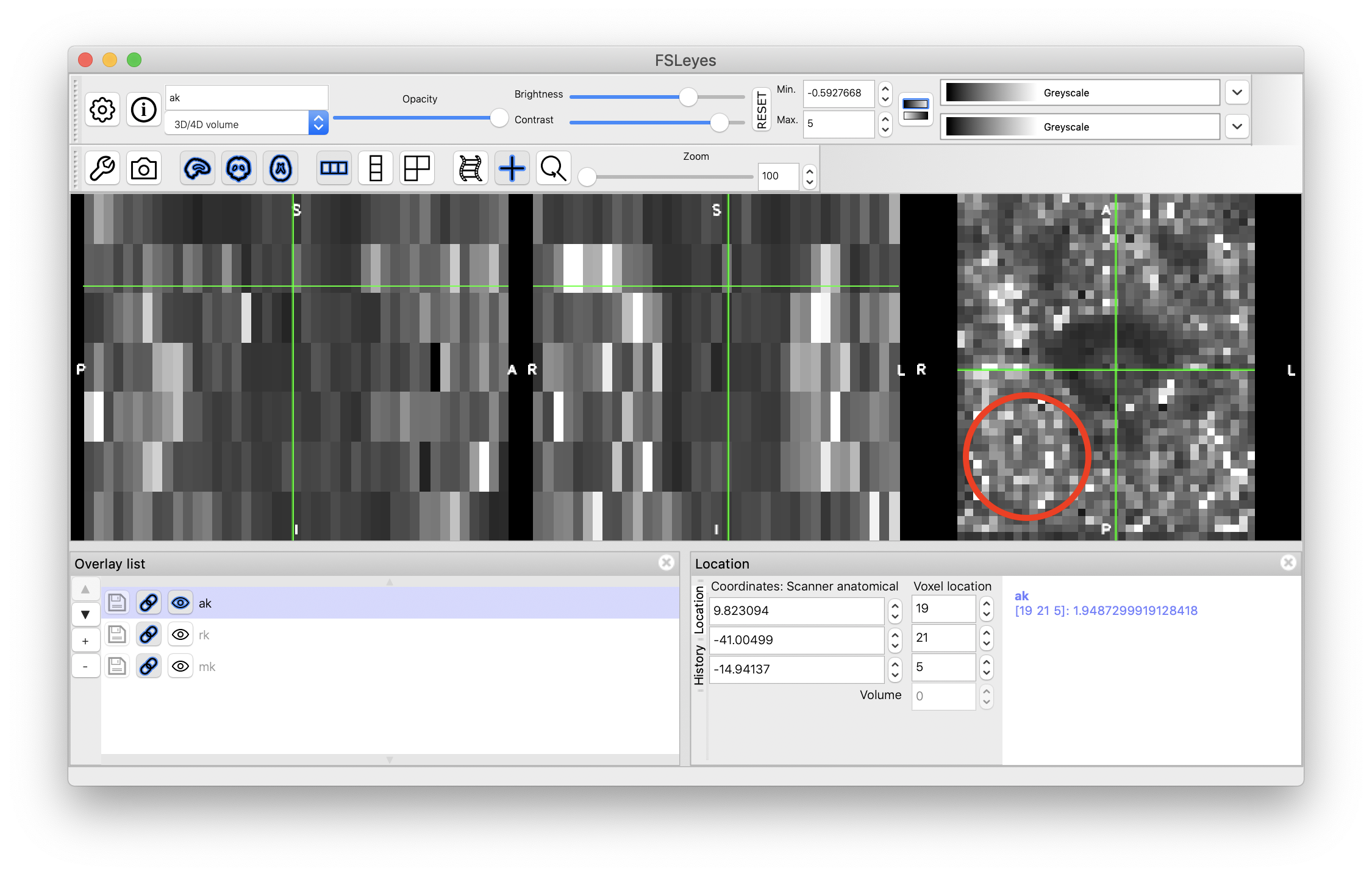
Hi,
If you are talking about the white/black pixels that you see in this red circle below, then please note that these pixel are not located in the spinal cord (probably in muscles/bone, hence diffusion signal is small, isotropic, and hence noisy). The spinal cord is located where the green cross is.