One more question. Is registration to PAM50 atlas suitable also for neonatal data? My data come from pediatric patients and I do not know if a more suitable atlas exist.
I mean the DKI metric’s value in left and right cortico-spinal tract are far different from each other and change every time I run sct_extract_metric.
values should not change everytime you run sct_extract_metric as the algorithm is deterministic (i.e. there is no randomization in this function). The file DWI_MK.csv you provided is very strange. Could you please upload the file mk.nii (compress it so it takes less space) and copy/paste the syntax you used so I can try to reproduce this behaviour?
One more question. Is registration to PAM50 atlas suitable also for neonatal data? My data come from pediatric patients and I do not know if a more suitable atlas exist.
so far, the PAM50 is the only spinal cord template/atlas compatible with SCT. So it is the only you can use so far. The registration should be tweaked as to allow sufficient deformation. Make sure you QC your registration results. For your information, we are working on adding a pediatric template on SCT, so this might be useful for you. More info here.
ok thank you, I attach the mk.nii.gz and the script where the last part is the sct_extract_metric command.
Rosellamk.nii.gz (40.7 KB)
script_kurtosis.sh (5.6 KB)
What do you mean with STD maps?
thanks, i would also need your folder label/atlas (please zip it before uploading it).
by STD map i meant the amplitude of the residues after fitting your data to the diffusion model. A few papers here:
ah, sorry i also need your label/template folder (because you are using vertebral levels for reference in your call to extract metrics).
For your information, in the meantime I tried running this command three times with the data you sent me, and the results are stable (contrary to what you observed):
sct_extract_metric -i mk.nii.gz -f atlas -l 4,5 -perslice 0 -o mk.csv -append 1
mk.csv (1.4 KB)
But let’s wait for your label/template folder, so i can reproduce exactly what you did.
Sorry for the delay but I was away from my office. Here is the label/template folder you asked for:
info_label.txt (723 Bytes)
PAM50_centerline.nii.gz (400 Bytes)
PAM50_cord.nii.gz (448 Bytes)
PAM50_csf.nii.gz (836 Bytes)
PAM50_gm.nii.gz (2.3 KB)
PAM50_label_body.nii.gz (326 Bytes)
PAM50_label_disc.nii.gz (300 Bytes)
PAM50_label_discPosterior.nii.gz (299 Bytes)
PAM50_levels.nii.gz (483 Bytes)
PAM50_levels_continuous.nii.gz (6.3 KB)
PAM50_spine.nii.gz (9.4 KB)
PAM50_t1.nii.gz (82.0 KB)
PAM50_t2.nii.gz (82.3 KB)
PAM50_t2s.nii.gz (82.0 KB)
PAM50_wm.nii.gz (5.1 KB)
Another question … I was asked to check whether the PAM50 template is suitable for segmentation of neonatal data through registering the PAM50 atlas to a 3DT1 image. Which is the best way to do so?
Thank you,
Rosella
Hi,
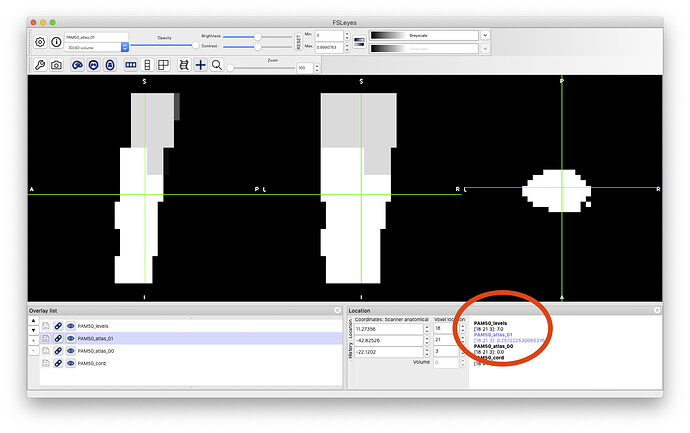
so, I looked at your vertebral level file, and it goes from 6 to 7, while you are asking to extract metrics between levels 2 and 5. So please, make sure your vertebral labeling is correct. See below:
Alternatively, if you wish to simply extract metrics throughout all your slices (and average across slices), you can run the following command:
sct_extract_metric -i mk.nii.gz -f atlas -l 4,5 -perslice 0 -o mk.csv -append 1
Another question … I was asked to check whether the PAM50 template is suitable for segmentation of neonatal data through registering the PAM50 atlas to a 3DT1 image. Which is the best way to do so?
Please see my answer here.
Best,
Julien
Thank you very much.
I will use the sct_register_to_template command in order to register the 3dT1 image to PAM 50 atlas.
Another precisation, by doing -perslice 0, do I compute the metric’s average value across all slices contained in the FA,MK,AK,RK etc image ?
THank you,
Rosella
yes, with -perslice 0, you compute the metric’s average value across all slices from the data specified in the flag -i.
I have just tried changing the sct_extract_metric command extended to all slices but the problem with my .csv file persists.
Here is the new mk.csv file
thank you ,
Rosella
DWI_MK.csv (819 Bytes)
wow, this is extremely bizarre. As if the file mk.nii is being corrupted when extracting metrics, which sounds strange. Could you please try to run this command below 5 times and upload your csv output:
sct_extract_metric -i mk.nii.gz -f label/template/PAM50_cord.nii.gz -perslice 0 -o mk_allcord.csv -append 1
Here it is!
Thank you for your patience but I do not actually know which the problem is!
Rosellamk_allcord.csv (813 Bytes)
ok, at least these values are stable. Now, could you try to run this 5x:
sct_extract_metric -i mk.nii.gz -f label/atlas -l 50 -perslice 0 -o mk_label50.csv -append 1
ok, we’re getting there  , now how about (run 5x):
, now how about (run 5x):
sct_extract_metric -i mk.nii.gz -f label/atlas -l 4,5 -perslice 0 -o mk_label4-5.csv -append 1
good. As you can see, values are unchanged across your trials, so you should be good to go