Dear all,
When I computing the csa of the same subject with T1 and T2 image, i got quite different result, it confused me.
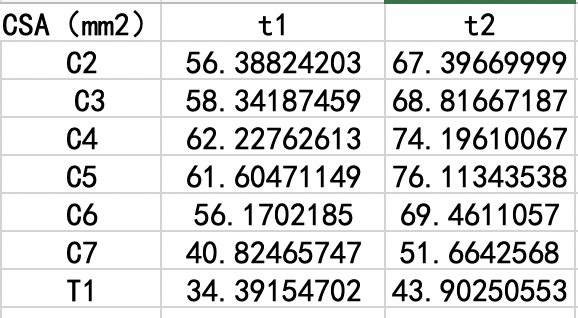
Almost each level of T1 is nearly 10 square millimeters less than T2.What’s wrong with it?
Thanks!
Dear all,
When I computing the csa of the same subject with T1 and T2 image, i got quite different result, it confused me.

Almost each level of T1 is nearly 10 square millimeters less than T2.What’s wrong with it?
Thanks!
Hi @wang,
This is totally normal, because the segmentation is driven by the image contrast at the CSF/cord interface, which differs across pulse sequence schemes and parameters. This is notably due to tissue at the interface, such as the pial matter, having different relaxation properties across contrasts. At the end, what matters for studies is to use the same contrast across patients & time point. More information about this:
An additional note: the under/over-segmentation on the deep learning approach (implemented in sct_deepseg_sc) is notably controlled by the threshold applied on the output prediction.
As of SCT 4.1.0 (link to pull request), we have introduced a threshold per contrast, which is optimized so that the CSA variation across contrasts is minimized. More information about the algorithm is available here.
I got it, thanks!