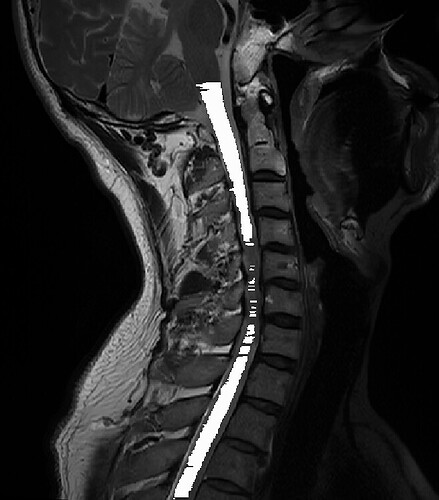
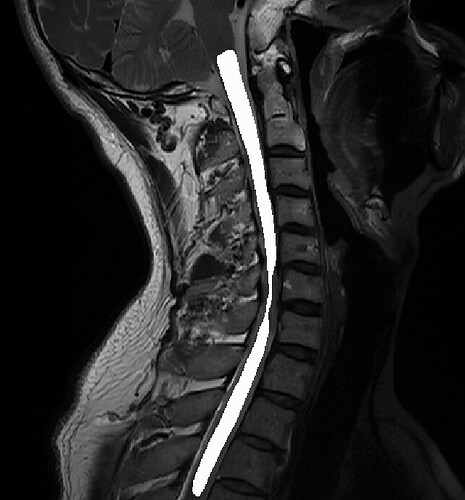
Dear SCT Team:
When I was doing spinal cord registration to the template, I encountered some of the data distortion in the sagittal position and coronal position, Could you provide me with the reasons behind this issue and possible solutions?

Hi @jie_Liu,
The issue could be caused by (among other things):
- straightening applied on axial data with thick slices
- too many degrees of freedom during registration
- issues with cord segmentation
I’m happy to look at it if you send me your image + the exact syntax you used (ideally, copy/paste the terminal output so we know exactly SCT’s version that was used)
Best,
Julien
Sorry for the late reply, here is my image file and the code I used (data2 I will reply again due to the limitation of upload file size)
data_and_code.rar (8.1 MB)
@jie_Liu, thank you for sharing your data and code-- however I realize that you sent multiple data, and multiple files, without instructions:
├── data
│ ├── data_1
│ │ ├── t2.nii.gz
│ │ └── t2s.nii.gz
│ ├── data_2
│ │ ├── t2.nii.gz
│ │ └── t2s.nii.gz
│ └── zeg_mask_0002.sh
├── readme.txt
└── seg_mask_spinal
├── c_seg.py
├── c_split.py
├── feature_sct.sh
├── seg.py
├── seg_add.py
├── seg_mask.sh
├── seg_mask_0001.sh
└── seg_mask_0318.sh
Moreover, the scripts have hard-coded absolute path.
I don’t have the time to figure out which script I should use, and which data the image above refers to.
In order for me to help you efficiently, please send me:
- ONE image (the one you show above)
- ONE script that I can simply run to reproduce your result
Also please, use ZIP instead of RAR
Dear jcohenadad, thank you for your reply.I’ve changed the path to a relative path so you can see it easily.Since the file is too large, I did a volume compression of the package.You can open the data terminal and type “bash zeg_mask_0002.sh” directly to run the script so that You can simply run to reproduce my result.
data_and_code.zip (3.2 MB)
data_and_code.z01 (5 MB)
data_and_code.z02 (5 MB)
data_and_code.z03 (5 MB)
data_and_code.z04 (5 MB)
And this is the ONE image (the one I show above).Since the file is too large that I can’t load it, I did a volume compression of the package.Decompress the zip file.
The_picture.zip (4.5 MB)
The_picture.z01 (5 MB)
The_picture.z02 (5 MB)
sorry, I cannot unzip your files-- i guess you can come back to RAR then…
I changed it back to rar.This is the ONE image (the one I show above).
The_picture.part1.rar (5 MB)
The_picture.part2.rar (5 MB)
The_picture.part3.rar (4.5 MB)
And this is the ONE script that you can simply run to reproduce my result.Thanks again.
data_and_code.part1.rar (5 MB)
data_and_code.part2.rar (5 MB)
data_and_code.part3.rar (5 MB)
data_and_code.part4.rar (5 MB)
data_and_code.part5.rar (3.2 MB)
just ONE single file please
Sorry, this file is too big and exceeds the upload limit of the website, and I can’t upload a compressed file directly.Can I have your email, I can send it to you directly by email.
use an cloud solution (eg dropbox, gdrive, etc.)
I used gdrive, can you check it?
https://drive.google.com/file/d/1RYF-0qj-GMOcB8d_MAzx7MXcOePMu_cn
Sorry, I’m using gdrive for the first time, I don’t have permission to modify, please use the new link.
https://drive.google.com/file/d/1RYF-0qj-GMOcB8d_MAzx7MXcOePMu_cn/view?usp=drivesdk
OK, I’ve looked at your data and script, and I was able to reproduce your results. A few comments:
-
The T2*w scan is 0.4 x 0.4 x 4.4 mm axial resolution. Straightening a highly anisotropy scan will always result in discontinuities.
-
The T2w scan is sagittal 3.3 x 0.5 x 0.5 mm resolution while the T2*w scan is 0.4 x 0.4 x 4.4 mm axial resolution. Trying to co-register the two will result in poor quality registration, especially because of the low R-L resolution of the sagittal (reference anatomical) scan. I advise against it, and instead use directly the T2*w scan to register to the PAM50, if you end up using the T2*w scan for subsequent analyses.
-
The segmentation on the T2w scan has a lot of discontinuities, leading to mis-registration issues (in addition to my previous point):
The new contrast-agnostic model (available as of V6.3 on
sct_deepseg) gives much better results, I recommend you use this method instead: -
L18 of your code, you used:
sct_register_multimodal -i t2.nii.gz -iseg t2_seg.nii.gz -d t2s.nii.gz -dseg t2s_seg.nii.gz -param step=1,type=seg,algo=centermass:step=2,type=seg,algo=bsplinesyn,slicewise=1,iter=3:step=3,type=im,algo=syn,slicewise=1,iter=1,metric=CC -owarp t22t2s/warp_t22t2s.nii.gz -owarpinv t22t2s/warp_t2s2t2.nii.gz -ofolder t22t2s/I would not use bsplinesyn algo for intra-subject intra-session co-registration. The centermass should suffice.
-
Your code masks out the spinal cord. I’m not sure what is the purpose of this step, given that the segmentation mask can be used to extract metrics inside the spinal cord.
Below is a suggestion to preform direct T2*w to PAM50 registration:
# Segment T2w
sct_deepseg -i t2.nii.gz -task seg_sc_contrast_agnostic -qc ~/qc_singleSubj
# Label vertebrae
sct_label_vertebrae -i t2.nii.gz -s t2_seg.nii.gz -c t2 -qc ~/qc_singleSubj
# Segment. T2*w
sct_deepseg -i t2s.nii.gz -task seg_sc_contrast_agnostic -qc ~/qc_singleSubj
# Bring vertebral levels to T2*w space (only use identity registration because of large axial slices, no need high precision)
sct_register_multimodal -i t2_seg_labeled.nii.gz -d t2s.nii.gz -dseg t2s_seg.nii.gz -identity 1 -x nn -qc ~/qc_singleSubj
# Create vertebral labels
sct_label_utils -i t2_seg_labeled_reg.nii.gz -vert-body 4,7 -o t2s_labels_vert_4-7.nii.gz
# Register T2*w to PAM50
sct_register_to_template -i t2s.nii.gz -s t2s_seg.nii.gz -l t2s_labels_vert_4-7.nii.gz -c t2 -qc ~/qc_singleSubj
Result below (toggle between PAM50 and registered T2*w):
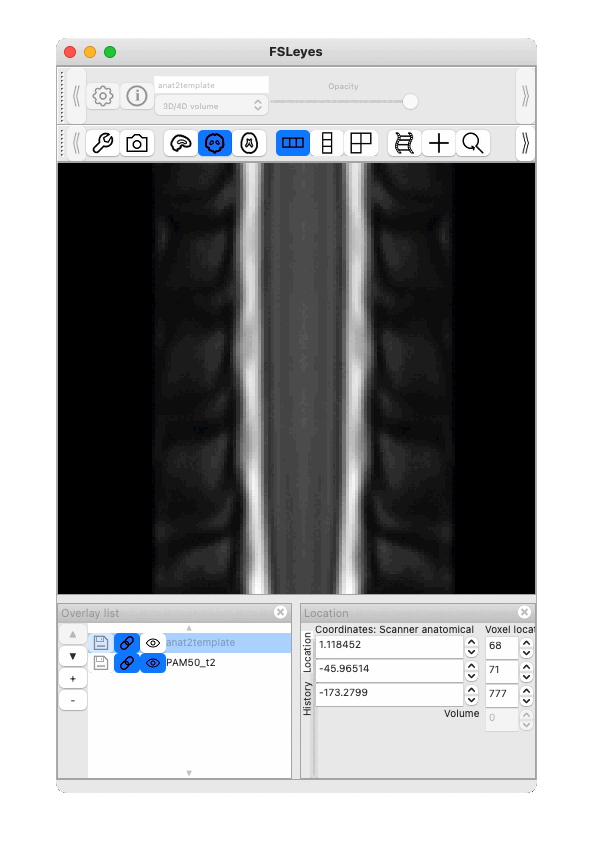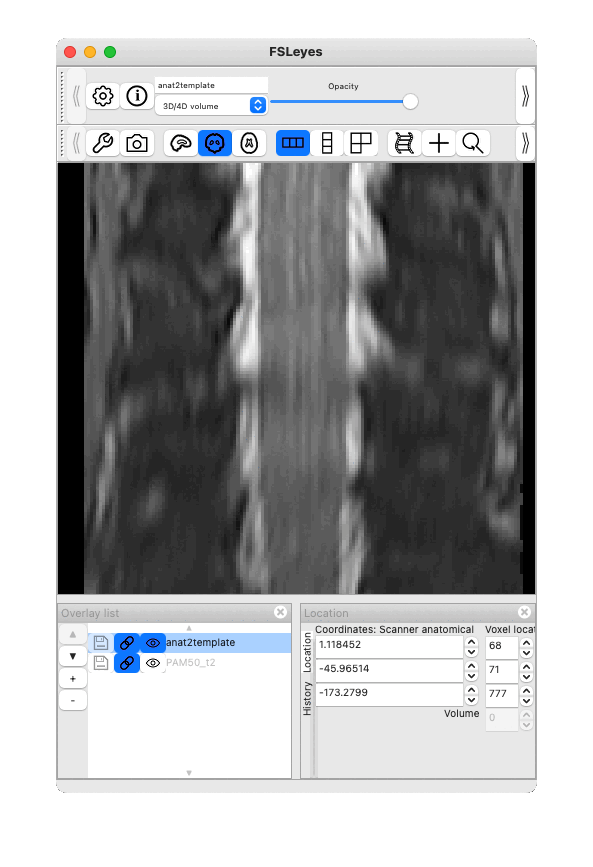
Thank you very much for your reply and your suggestions!