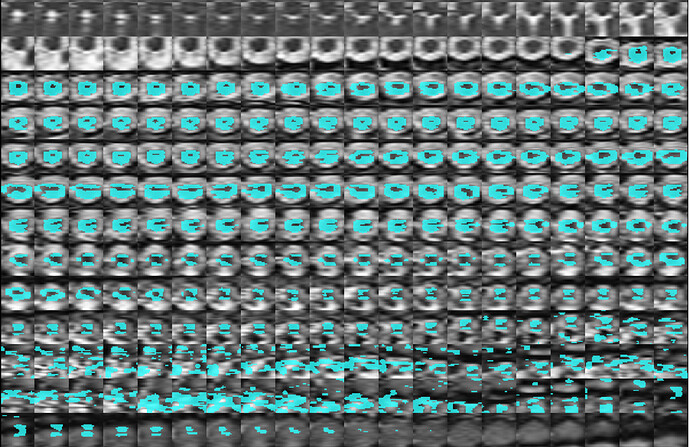
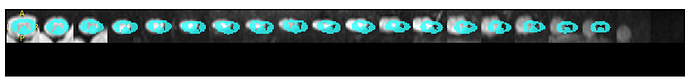
Dear @jcohenadad and SCT Team!
After running this command:
“sct_warp_template -d t2.nii.gz -w warp_template2anat.nii.gz -a 0 -qc”
my qc output is as follows:
This is the final product combining T2 anatomical with the DTI:
I am having some trouble correcting what I believe might be a registration error? Any assistance would be greatly appreciated!
All the best,
Adriana
hi Adriana,
i suspect two (related) issues:
- the anatomical T2 scan you are using for segmentation (and template registration) is sagittal with thick slices, which is not recommended (low right-left resolution)
- poor segmentation on the anatomical T2 image, which induced poor template registration (as it is using the segmentation).
Because of 1, i would suggest to only use the T2w scan for automatic vertebral labeling, and register the template directly on the diffusion scans (using flag -ref subject). An example is available in the latest SCT course.
Cheers,
Julien
Thank you so much for the reply!
How would you suggest Are there any methods by which I can improve the segmentation on the anatomical T2 image?
Thanks!
Adriana
the problem is not the method, it is the data. Thick sagittal slices are not recommended for spinal cord segmentation. How thick are your slices? (and what gap?)
Thank you that clarifies things. The slices are 5.5 mm thick.
Thank you that clarifies things. The slices are 5.5 mm thick.
Ok, given that the cord is about 8mm thick in the right-left direction, there is no way you could get a proper cord segmentation with such high thickness: the cord is covered by only 2 slices (!).
Is there a way to use a template anatomical spinal cord, and register my subject DTIs to that, rather than to the subject anatomical T2?
yes, as i mentioned above, you can register the template directly with the diffusion scans (using flag -ref subject ). An example is available in the latest SCT course, or you can also see some examples here, notably this one.
Thank you!
I appear to be having trouble registering to the diffusion scan as it is not a 3D volume? Perhaps I am using the wrong image file. This is what I have done:
sct_register_to_template -i dmri_crop.nii.gz -s t2_seg.nii.gz -l t2_labels_vert.nii.gz -ref subject -c t2 -param step=1,type=seg,algo=centermassrot,smooth=0:step=2,type=seg,algo=columnwise,smooth=0,smoothWarpXY=2 -qc ~/Documents/Research/CP_SDR/SCT_data/DTI_04/
indeed, there are probably some confusions about what warping field you used for what data. The easiest way to debug, at this point, would be for you to share the data along with the analysis script. Please avoid absolute path-- i need to be able to reproduce your analysis without touching the script.
then, i will fix the script and upload it here.
Thank you very much! I will share both via email.
Hi,
Here is a script that works:
#!/bin/bash/
# t2
# ==============================================================================
# Segment spinal cord
sct_deepseg_sc -i t2.nii.gz -c t2 -qc qc
# Manual initialization of disc C2-C3
sct_label_utils -i t2.nii.gz -create-viewer 3 -o label_disc_c2c3.nii.gz
# Vertebral labeling
# Tips: given that the spine is smaller than in average adults, we used -scale-dist to adapt the scaling and disc detection.
sct_label_vertebrae -i t2.nii.gz -s t2_seg.nii.gz -c t2 -initlabel label_disc_c2c3.nii.gz -scale-dist 0.8 -qc qc
# Create labels at in the cord at C2 and C5 mid-vertebral levels
sct_label_utils -i t2_seg_labeled.nii.gz -vert-body 2,5 -o t2_labels_vert.nii.gz
# dwi
# ==============================================================================
# Bring t2 segmentation in dmri space to create mask (no optimization)
sct_maths -i dmri.nii.gz -mean t -o dmri_mean.nii.gz
sct_register_multimodal -i t2_seg.nii.gz -d dmri_mean.nii.gz -identity 1 -x linear
# Create Gaussian mask to increase accuracy of motion correction
sct_create_mask -i dmri_mean.nii.gz -p centerline,t2_seg_reg.nii.gz -f gaussian -size 35mm
# Motion correction
# Tips: if data have very low SNR you can increase the number of successive images that are averaged into group with "-g". Also see: sct_dmri_moco -h
sct_dmri_moco -i dmri.nii.gz -bvec bvecs.bvec -m mask_dmri_mean.nii.gz
# Segment cord on the DWI data
sct_deepseg_sc -i dmri_moco_dwi_mean.nii.gz -c dwi -qc qc
# Register to template directly to the dmri data, given the thick slices on the t2 scan
sct_register_to_template -i dmri_moco_dwi_mean.nii.gz -s dmri_moco_dwi_mean_seg.nii.gz -l t2_labels_vert.nii.gz -ref subject -c t1 -param step=1,type=seg,algo=centermassrot,smooth=0:step=2,type=seg,algo=bsplinesyn,smooth=0,iter=5,slicewise=1:step=3,type=im,algo=bsplinesyn,slicewise=1,iter=3 -qc qc
# Rename warping fields for clarity
mv warp_template2anat.nii.gz warp_template2dmri.nii.gz
mv warp_anat2template.nii.gz warp_dmri2template.nii.gz
# Warp template
sct_warp_template -d dmri_moco_dwi_mean.nii.gz -w warp_template2dmri.nii.gz -qc qc
# Compute DTI metrics
sct_dmri_compute_dti -i dmri_moco.nii.gz -bval bvals.txt -bvec bvecs.txt
# Compute FA in the white matter between C1 and C5
sct_extract_metric -i dti_FA.nii.gz -l 51 -v 1:5 -method map -o fa_in_wm.csv
Note: you did not share the bvals file so I cannot test the script entirely. But you can easily figure out how to adapt the script (just update the name of the bvals file).
Thank you very much for your assistance!
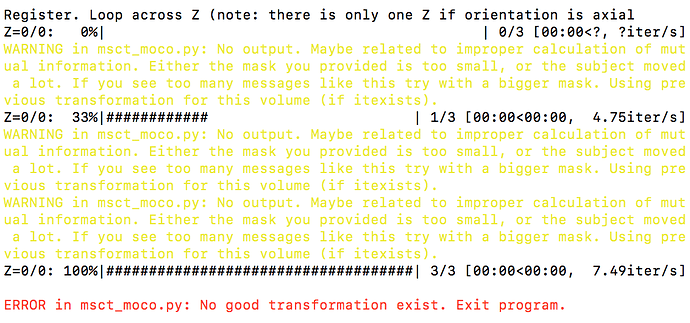
Hello! I seem to be getting an error at the motion correction step:
are you using the same data you sent me? If so, what is the output of: sct_version?
Yes I am! I am using version 4.1.0
can you please install the latest version from git’s master branch? Here are the instructions.
I downloaded the latest version and it works! Thank you!
1 Like
Hello @jcohenadad!
Following up, I seem to be getting a new error. When I run sct_dmri_moco -i dmri.nii.gz -bvec bvecs.bvec -m mask_dmri_mean.nii.gz, I get this error:
RunError(output)
sct_utils.RunError: Warning: bad time point - too little intensity variation0 0
libc++abi.dylib: terminating with uncaught exception of type itk::ExceptionObject: /Users/runner/work/ANTs/ANTs/antsbin/staging/include/ITK-5.1/itkImageBase.hxx:177:
itk::ERROR: Image(0x7fe7de5bee60):
A spacing of 0 is not allowed: Spacing is [0, 0]
Once again, I would really appreciate your input! Thank you so much in advance