Hi @jcohenadad,
I ran my updated scripts on 32 datasets and they worked well on 31. The SCT code was, however, unable to register one of the datasets to the template. This particular MR image looks good and the spinal cord is segmented properly, so I am not sure why it is unable to warp to the template. The only user interaction is manually identifying C2/C3; I re-identified this point and got the same output errors (pasted below). Do you have an idea of what might be happening?
Thanks!
-Rob
Warping field generated: tmp.curve2straight.nii.gz
Warping field generated: tmp.straight2curve.nii.gz
Apply transformation to input image…
e[94m/Users/rob/spinalcordtoolbox/bin/isct_antsApplyTransforms -d 3 -r tmp.centerline_pad_crop.nii.gz -i data.nii -o tmp.anat_rigid_warp.nii.gz -t tmp.curve2straight.nii.gz -n ‘BSpline[3]’ # in /private/var/folders/c_/x6z5ffcj2wbc0h68_g1h0l_h0000gn/T/sct-20210225055438.266275-straighten_spinalcord-shrsqloge[0m
Generate output files…
File created: ./warp_curve2straight.nii.gz
File created: ./warp_straight2curve.nii.gz
e[94mcp /var/folders/c_/x6z5ffcj2wbc0h68_g1h0l_h0000gn/T/sct-20210225055438.266275-straighten_spinalcord-shrsqlog/tmp.anat_rigid_warp.nii.gz ./straight_ref.nii.gze[0m
File created: ./data_straight.nii
Remove temporary files…
e[94mrm -rf /var/folders/c_/x6z5ffcj2wbc0h68_g1h0l_h0000gn/T/sct-20210225055438.266275-straighten_spinalcord-shrsqloge[0m
e[0m
Finished! Elapsed time: 366 se[0m
e[0m
Resample to 0.5mm isotropic…e[0m
e[94msct_resample -i data_straight.nii -mm 0.5x0.5x0.5 -x linear -o data_straightr.nii # in /private/var/folders/c_/x6z5ffcj2wbc0h68_g1h0l_h0000gn/T/sct-20210225055434.496959-label_vertebrae-ow9z9cvqe[0m
e[0m
Apply straightening to segmentation…e[0m
e[94m/Users/rob/spinalcordtoolbox/bin/isct_antsApplyTransforms -d 3 -i segmentation.nii -r data_straightr.nii -t warp_curve2straight.nii.gz -o segmentation_straight.nii -n Linear # in /private/var/folders/c_/x6z5ffcj2wbc0h68_g1h0l_h0000gn/T/sct-20210225055434.496959-label_vertebrae-ow9z9cvqe[0m
e[33mFile /private/var/folders/c_/x6z5ffcj2wbc0h68_g1h0l_h0000gn/T/sct-20210225055434.496959-label_vertebrae-ow9z9cvq/segmentation_straight.nii already exists. Will overwrite it.e[0m
e[0m
Create label to identify disc…e[0m
e[33mFile /var/folders/c_/x6z5ffcj2wbc0h68_g1h0l_h0000gn/T/sct-20210225055434.496959-label_vertebrae-ow9z9cvq/labelz.nii.gz already exists. Will overwrite it.e[0m
e[0m
And apply straightening to label…e[0m
e[94m/Users/rob/spinalcordtoolbox/bin/isct_antsApplyTransforms -d 3 -i labelz.nii.gz -r data_straightr.nii -t warp_curve2straight.nii.gz -o labelz_straight.nii.gz -n NearestNeighbor # in /private/var/folders/c_/x6z5ffcj2wbc0h68_g1h0l_h0000gn/T/sct-20210225055434.496959-label_vertebrae-ow9z9cvqe[0m
e[0m
Get z and disc values from straight label…e[0m
e[0m… [365, 3]e[0m
Look for template…
Path template: /Users/rob/SCT/data/PAM50
Open template and vertebral levels…
Disc values from template: [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19]
Z-values for each disc: [963, 938, 907, 870, 833, 800, 769, 735, 692, 646, 600, 551, 500, 449, 396, 342, 289, 231, 168, 104, 79]
Distances between discs (in voxel): [25.0, 31.0, 37.0, 37.0, 33.0, 31.0, 34.0, 43.0, 46.0, 46.0, 49.0, 51.0, 51.0, 53.0, 54.0, 53.0, 58.0, 63.0, 64.0, 25.0]
Detect intervertebral discs…
Current disc: 2 (z=365). Direction: superior
… Peak found: z=-1 (correlation = 0.5806519076174395)
Current disc: 1 (z=395). Direction: superior
… correcting factor: 1.0
… Switching to inferior direction.
Current disc: 3 (z=328). Direction: inferior
… Peak found: z=-1 (correlation = 0.6659052966517715)
… correcting factor: 1.0
Current disc: 4 (z=290). Direction: inferior
… Peak found: z=1 (correlation = 0.6105950224899827)
… correcting factor: 0.990990990990991
Current disc: 5 (z=258). Direction: inferior
… Peak found: z=0 (correlation = 0.44450624761912827)
… correcting factor: 0.9932432432432432
Current disc: 6 (z=227). Direction: inferior
… Peak found: z=-2 (correlation = 0.5430550901155323)
… correcting factor: 1.007497820401046
Current disc: 7 (z=191). Direction: inferior
… Peak found: z=2 (correlation = 0.43807046883648043)
… correcting factor: 0.996444262098911
Current disc: 8 (z=150). Direction: inferior
… Peak found: z=1 (correlation = 0.554691318759119)
… correcting factor: 0.9936299655199968
Current disc: 9 (z=105). Direction: inferior
… Peak found: z=1 (correlation = 0.4752640839639358)
… correcting factor: 0.9917088285256495
Current disc: 10 (z=60). Direction: inferior
… Peak found: z=-1 (correlation = 0.4791790829143412)
… correcting factor: 0.9950455287377754
Current disc: 11 (z=10). Direction: inferior
… Peak found: z=5 (correlation = 0.5583133338704257)
… correcting factor: 0.9853368942313449
Adding top disc based on adjusted template distance: #0
… approximate distance: 25
Traceback (most recent call last):
File “/Users/rob/spinalcordtoolbox/spinalcordtoolbox/scripts/sct_label_vertebrae.py”, line 491, in
main(sys.argv[1:])
File “/Users/rob/spinalcordtoolbox/spinalcordtoolbox/scripts/sct_label_vertebrae.py”, line 425, in main
verbose=verbose, path_template=path_template, path_output=path_output, scale_dist=scale_dist)
File “/Users/rob/spinalcordtoolbox/spinalcordtoolbox/vertebrae/core.py”, line 251, in vertebral_detection
label_discs(fname_seg, list_disc_z, list_disc_value, verbose=verbose)
File “/Users/rob/spinalcordtoolbox/spinalcordtoolbox/vertebrae/core.py”, line 522, in label_discs
cx, cy = [int(x) for x in np.round(center_of_mass(slices)).tolist()]
File “/Users/rob/spinalcordtoolbox/spinalcordtoolbox/vertebrae/core.py”, line 259, in center_of_mass
raise ValueError(“Array has no mass”)
ValueError: Array has no mass
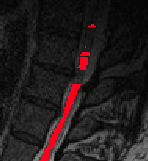
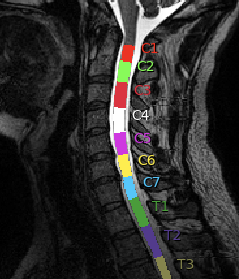
 You had several comments or questions so I’ll do my best to address each of them.
You had several comments or questions so I’ll do my best to address each of them.