Hello
When trying to register a template to the dMRI, the following error message is occurring:
“ValueError: Displacement field in warp_template2t2s.nii is invalid: should be encoded in a 5D file with vector intent code (see https://nifti.nimh.nih.gov/pub/dist/src/niftilib/nifti1.h)”
Here are the commands we used to get the warp_template2t2s file:
cd …/t2s
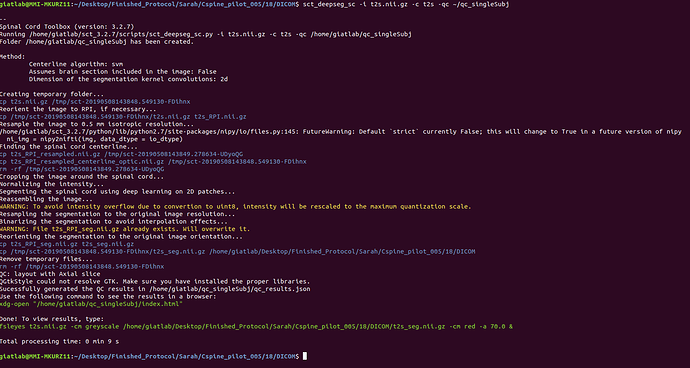
sct_deepseg_sc -i t2s.nii.gz -c t2s -qc ~/qc_singleSubj
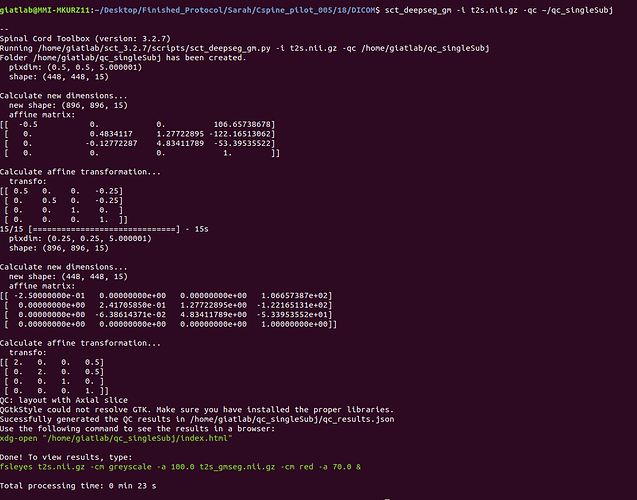
sct_deepseg_gm -i t2s.nii.gz -qc ~/qc_singleSubj
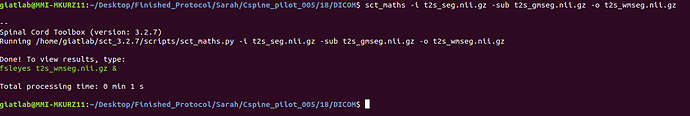
sct_maths -i t2s_seg.nii.gz -sub t2s_gmseg.nii.gz -o t2s_wmseg.nii.gz
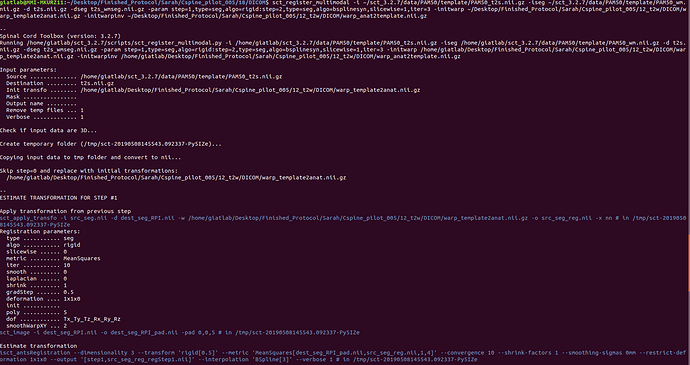
sct_register_multimodal -i ${SCT_DIR}/data/PAM50/template/PAM50_t2s.nii.gz -iseg $
{SCT_DIR}/data/PAM50/template/PAM50_wm.nii.gz -d t2s.nii.gz -dseg t2s_wmseg.nii.gz -param
step=1,type=seg,algo=rigid:step=2,type=seg,algo=bsplinesyn,slicewise=1,iter=3 -initwarp …/t2/
warp_template2anat.nii.gz -initwarpinv …/t2/warp_anat2template.nii.gz -qc ~/qc_singleSubj
mv warp_PAM50_t2s2t2s.nii.gz warp_template2t2s.nii.gz
mv warp_t2s2PAM50_t2s.nii.gz warp_t2s2template.nii.gz
sct_warp_template -d t2s.nii.gz -w warp_template2t2s.nii.gz -qc ~/qc_singleSubj
Thank you.
Hi,
if you have FSL, what is the output of fslhd t2s.nii.gz? And fslhd warp_template2t2s.nii.gz
Hello,
fslhd t2s.nii.gz output:
sizeof_hdr 348
data_type INT16
dim0 3
dim1 448
dim2 448
dim3 15
dim4 1
dim5 1
dim6 1
dim7 1
vox_units mm
time_units s
datatype 4
nbyper 2
bitpix 16
pixdim0 0.000000
pixdim1 0.500000
pixdim2 0.500000
pixdim3 5.000001
pixdim4 0.600000
pixdim5 1.000000
pixdim6 1.000000
pixdim7 35393.449219
vox_offset 352
cal_max 0.0000
cal_min 0.0000
scl_slope 1.000000
scl_inter 0.000000
phase_dim 0
freq_dim 0
slice_dim 3
slice_name alternating_increasing
slice_code 3
slice_start 0
slice_end 14
slice_duration 0.000000
time_offset 0.000000
intent Unknown
intent_code 0
intent_name
intent_p1 0.000000
intent_p2 0.000000
intent_p3 0.000000
qform_name Scanner Anat
qform_code 1
qto_xyz:1 -0.500000 0.000000 -0.000000 106.657387
qto_xyz:2 0.000000 0.483412 1.277229 -122.165131
qto_xyz:3 -0.000000 -0.127723 4.834118 -53.395355
qto_xyz:4 0.000000 0.000000 0.000000 1.000000
qform_xorient Right-to-Left
qform_yorient Posterior-to-Anterior
qform_zorient Inferior-to-Superior
sform_name Scanner Anat
sform_code 1
sto_xyz:1 -0.500000 0.000000 0.000000 106.657387
sto_xyz:2 0.000000 0.483412 1.277229 -122.165131
sto_xyz:3 0.000000 -0.127723 4.834118 -53.395355
sto_xyz:4 0.000000 0.000000 0.000000 1.000000
sform_xorient Right-to-Left
sform_yorient Posterior-to-Anterior
sform_zorient Inferior-to-Superior
file_type NIFTI-1+
file_code 1
descrip TE=14;sec=35393.4475;phaseDir=+
aux_file
Thanks. Could you also send us the output of
fslhd warp_template2t2s.nii.gz
Hello,
fslhd warp_template2t2s.nii.gz output:
sizeof_hdr 348
data_type FLOAT64
dim0 3
dim1 448
dim2 448
dim3 15
dim4 1
dim5 1
dim6 1
dim7 1
vox_units mm
time_units Unknown
datatype 64
nbyper 8
bitpix 64
pixdim0 0.000000
pixdim1 0.500000
pixdim2 0.500000
pixdim3 5.000001
pixdim4 1.000000
pixdim5 1.000000
pixdim6 1.000000
pixdim7 1.000000
vox_offset 352
cal_max 0.0000
cal_min 0.0000
scl_slope 1.000000
scl_inter 0.000000
phase_dim 0
freq_dim 0
slice_dim 0
slice_name Unknown
slice_code 0
slice_start 0
slice_end 0
slice_duration 0.000000
time_offset 0.000000
intent Unknown
intent_code 0
intent_name
intent_p1 0.000000
intent_p2 0.000000
intent_p3 0.000000
qform_name Aligned Anat
qform_code 2
qto_xyz:1 -0.500000 0.000000 -0.000000 106.657387
qto_xyz:2 -0.000000 0.483412 1.277229 -122.165131
qto_xyz:3 0.000000 -0.127723 4.834118 -53.395355
qto_xyz:4 0.000000 0.000000 0.000000 1.000000
qform_xorient Right-to-Left
qform_yorient Posterior-to-Anterior
qform_zorient Inferior-to-Superior
sform_name Scanner Anat
sform_code 1
sto_xyz:1 -0.500000 -0.000000 -0.000000 106.657387
sto_xyz:2 -0.000000 0.483412 1.277229 -122.165131
sto_xyz:3 0.000000 -0.127723 4.834118 -53.395355
sto_xyz:4 0.000000 0.000000 0.000000 1.000000
sform_xorient Right-to-Left
sform_yorient Posterior-to-Anterior
sform_zorient Inferior-to-Superior
file_type NIFTI-1+
file_code 1
descrip
aux_file
OK, for some reasons, the file warp_template2t2s.nii.gz is an image and not a warping field.
Could you please re-run all the commands that you listed in your original message above, including the command that gives you the error, and copy/paste the entire terminal output, so I can spot what went wrong?
Cheers,
Julien
Hi Julien,
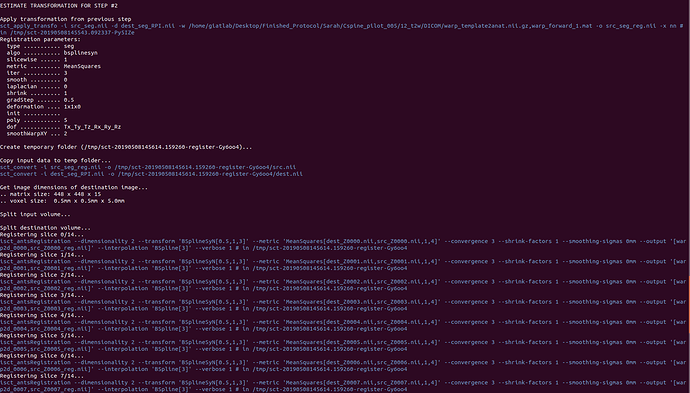
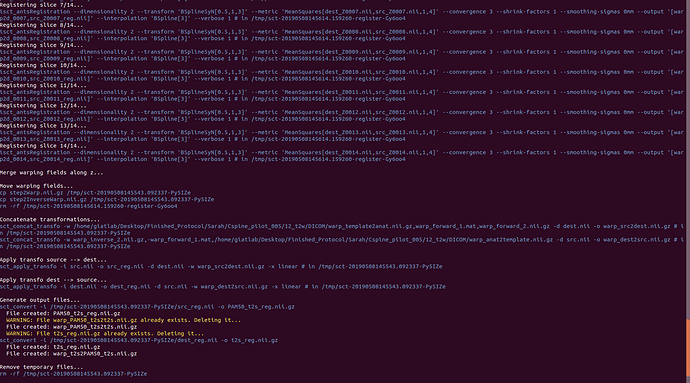
Attached are screenshots of the output from the commands of the original message.
hi,
sorry but i am not able to retrace the entire processing workflow from these screenshots (e.g., the list of commands that generated the input of -initwarp is not shown).
the easiest for me would be to run the list of commands on my end, to be able to spot the problem. If
you are able to share (privately) the data needed to re-create your error, please send me your full list of commands (incl. registration to the template from the t2w scan), and i can have a look at it.
best,
Julien