Hey there,
I am new to the whole SCT topic & coding in general, so sorry if i make mistakes that are easy to fix, I am using this software for an academic paper.
I am using the following commands:
sct_propseg -i “T2 FILENAME” -c t2 -init-centerline viewer
sct_label_utils -i “T2 FILENAME” -create-viewer 2,5 -o labels_vert.nii.gz
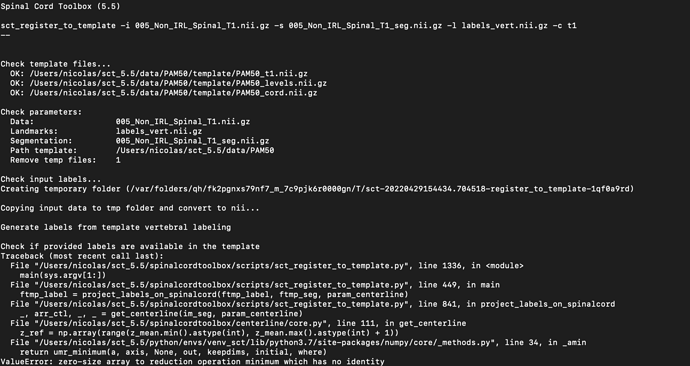
sct_register_to_template -i “T2 FILENAME” -s “T2 SEG FILE” -l labels_vert.nii.gz -c t2
sct_warp_template -d anat2template.nii.gz -w warp_anat2template.nii.gz -a 0
the first two steps are never a problem. However sometimes (so not always and not following a strict pattern) the third step, the register_to_template doesn’t work, I get the error code “ValueError: zero-size array to reduction operation minimum which has no identity”. It is completely random whether it happens or not, i always do the same procedure.
My System information are as follows:
SCT info:
- version: 5.5
- path: /Users/nicolas/sct_5.5
OS: osx (Darwin-21.4.0-x86_64-i386-64bit)
CPU cores: Available: 4, Used by ITK functions: 4
RAM: Total: 8192MB, Used: 4834MB, Available: 2795MB
Check Python executable…[OK]
Using bundled python 3.7.11 (default, Jul 27 2021, 07:03:16)
[Clang 10.0.0 ] at /Users/nicolas/sct_5.5/python/envs/venv_sct/bin/python
Check if data are installed…[OK]
Check if colored is installed…[OK] (1.4.3)
Check if dipy is installed…[OK] (1.4.1)
Check if h5py is installed…[OK] (2.10.0)
Check if Keras (2.3.1) is installed…[OK] (2.3.1)
Check if ivadomed is installed…[OK] (2.9.2)
Check if matplotlib is installed…[OK] (3.5.1)
Check if nibabel is installed…[OK] (3.2.1)
Check if numpy is installed…[OK] (1.18.5)
Check if onnxruntime is installed…[OK] (1.7.0)
Check if pandas is installed…[OK] (1.3.5)
Check if psutil is installed…[OK] (5.9.0)
Check if pyqt5 (5.11.3) is installed…[OK] (5.11.3)
Check if pytest is installed…[OK] (6.2.5)
Check if pytest-cov is installed…[OK] (3.0.0)
Check if raven is installed…[OK]
Check if requests is installed…[OK] (2.27.1)
Check if requirements-parser is installed…[OK]
Check if scipy is installed…[OK] (1.7.3)
Check if scikit-image is installed…[OK] (0.19.1)
Check if scikit-learn is installed…[OK] (1.0.2)
Check if tensorflow is installed…[OK] (1.15.5)
Check if torch (1.5.0) is installed…[OK] (1.5.0)
Check if torchvision (0.6.0) is installed…[OK] (0.6.0)
Check if xlwt is installed…[OK] (1.3.0)
Check if tqdm is installed…[OK] (4.62.3)
Check if transforms3d is installed…[OK] (0.3.1)
Check if urllib3 is installed…[OK] (1.26.8)
Check if pytest_console_scripts is installed…[OK]
Check if wquantiles is installed…[OK] (0.4)
Check if spinalcordtoolbox is installed…[OK]
Check ANTs compatibility with OS …[OK]
Check PropSeg compatibility with OS …[OK]
Check if figure can be opened with matplotlib…[OK] (Using GUI backend: ‘MacOSX’)
Check if figure can be opened with PyQt…[OK]
Check FSLeyes version…Command not found. If you installed FSLeyes as part of FSL package, please check that FSL is included in $PATH variable. If you installed FSLeyes using conda environment, make sure that the environment is activated. If you do not have FSLeyes installed, consider its installation to easily visualize processing outputs and/or to use SCT within FSLeyes. More info at: FSLeyes Integration - Spinal Cord Toolbox documentation
I can give more information as needed and I would be really grateful if somebody finds an answer to my struggles!
Kindest Regards,
Nicolas