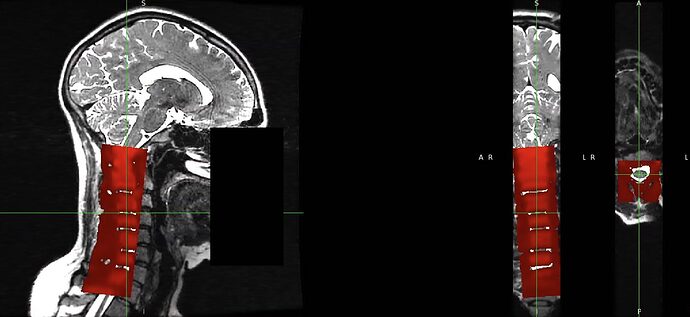
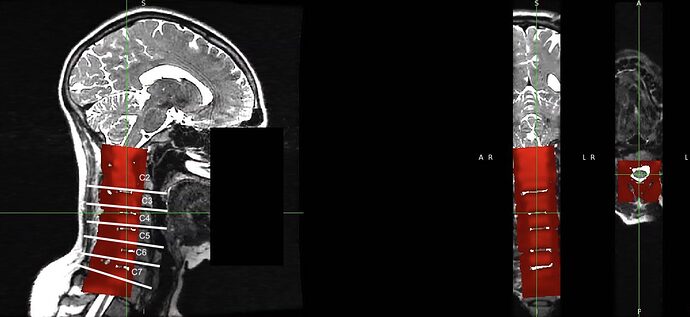
Hi Julien/SCT team,
Aim: we’d like to register EPI data to high-res 3D T2weighted scans (e.g. SPACE) of the same subject. EPI and T2w acquired in the same session, but cannot assume they haven’t moved between acquisitions.
Reason: would like to register to template using this intermediate (helper) image (the T2w)
i.e.
(step 1) EPI → T2w (sct_register_multimodal)
(step 2) T2w → PAM50_T2s (sct_register_to_template)
by concatenating the transformations for steps (1) and (2), provide an initial warp that puts the PAM50_T2s in much closer alignment to the EPI - giving it the best chance to work.
(step 3) EPI → PAM50_T2s (using initial warp concatenated from steps (1) & (2)).
Problems:
(1) EPI data have thick slices. Unfortunately no way around this…
(2) We have to manually edit the cord mask on the EPI - again, no way around this with the current data quality
(3) Registration with the following command (step 1) gives apparently good result, but there is a mismatch in the location of intervertebral discs.
sct_register_multimodal -i T2w.nii.gz -iseg T2w_seg.nii.gz -d EPI_moco_mean.nii.gz -dseg EPI_moco_mean_seg.nii.gz -param step=1,type=seg,algo=centermass:step=2,type=im,algo=syn,metric=MI,slicewise=1,smooth=0,iter=3
To check on the rostro-caudal alignment I took the original EPI and rubbed out the signal surrounding the cord on each slice containing a disc (clearly this step is subjective).
I then applied the warp field estimated with the step above to this “disc erased” image to bring into the space of the T2w.
It would be great if we could use something similar to the -ldisc argument found in sct_register_to_template, but would allow the user to specify a disc labelling for the input and destination for sct_register_multimodal.
I know it’s possible to go straight from EPI to template using an -ldisc created on the EPI (with sct_register_to_template), but in our experience having an initwarp appears to help a lot (which would ideally be the concatenation of warps resulting from steps 1 & 2, see above) - we use sct_register_multimodal for this step as this “allows” the use of initwarp and initwarpinv.
Would appreciate any comments, suggestions on how to achieve this.
Best wishes,
Jon