Me again,
Using dev version, after 4.2.1
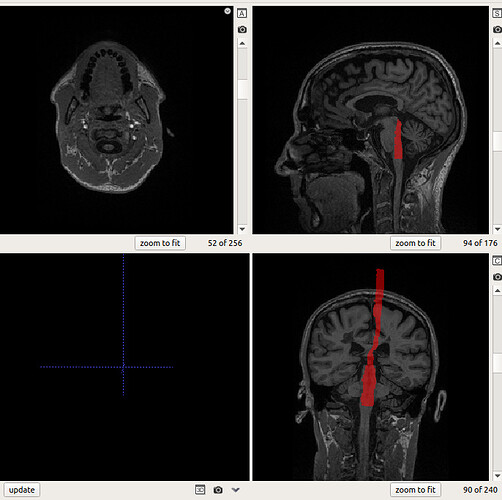
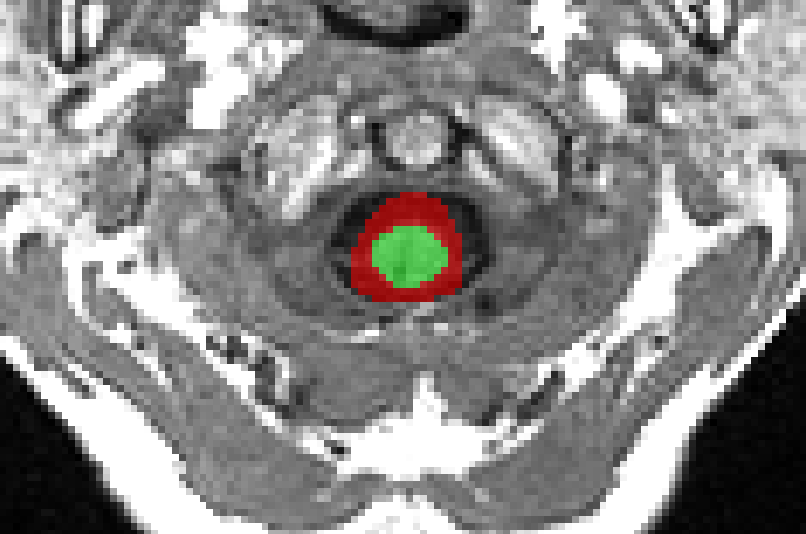
Trying to obtain the CSF segmentation using sct_propseg, we encounter the following issues.
The code gives an error at some point but is able to extract images [EXPLAINED BELOW]
extop@:TT1$ sct_propseg -i T1.nii -c t1 -CSF
--
Spinal Cord Toolbox (dev)
Create temporary folder (/tmp/sct-20200224122453.203680-label_vertebrae-r4uo4yt7)...
Remove temporary files...
rm -rf /tmp/sct-20200224122453.204806-asblo5d9
/home/extop/sct_dev/bin/isct_propseg -t t1 -o ./ -verbose -CSF -i '/T1.nii' -init-centerline /tmp/sct-20200224122453.203680-label_vertebrae-r4uo4yt7/centerline_optic.nii.gz -centerline-binary
Check consistency of segmentation...
Create temporary folder (/tmp/sct-20200224122539.013849-propseg-46lybd2z)...
tmp.segmentation.nii.gz
tmp.centerline.nii.gz
Get data dimensions...
usage: sct_image -i <file> [<file> ...] [-h] [-o <file>] [-pad <list>]
[-pad-asym <list>] [-split {x,y,z,t}] [-concat {x,y,z,t}]
[-remove-vol <list>] [-keep-vol <list>]
[-type {uint8,int16,int32,float32,complex64,float64,int8,uint16,uint32,int64,uint64}]
[-copy-header <file>] [-getorient]
[-setorient {RIP,LIP,RSP,LSP,RIA,LIA,RSA,LSA,IRP,ILP,SRP,SLP,IRA,ILA,SRA,SLA,RPI,LPI,RAI,LAI,RPS,LPS,RAS,LAS,PRI,PLI,ARI,ALI,PRS,PLS,ARS,ALS,IPR,SPR,IAR,SAR,IPL,SPL,IAL,SAL,PIR,PSR,AIR,ASR,PIL,PSL,AIL,ASL}]
[-setorient-data {RIP,LIP,RSP,LSP,RIA,LIA,RSA,LSA,IRP,ILP,SRP,SLP,IRA,ILA,SRA,SLA,RPI,LPI,RAI,LAI,RPS,LPS,RAS,LAS,PRI,PLI,ARI,ALI,PRS,PLS,ARS,ALS,IPR,SPR,IAR,SAR,IPL,SPL,IAL,SAL,PIR,PSR,AIR,ASR,PIL,PSL,AIL,ASL}]
[-mcs] [-omc] [-display-warp] [-v {0,1,2}]
sct_image: error: unrecognized arguments: EXT/T1.nii
but the CSF_segmentation exists in all slices and is centered wrongly.
I believe to have found the origin of the first error. We are running the analysis in a mounted unit and i think the program loses the correct path.
It should not be EXT but the name of the unit.
EDIT: Probably it origins with the name of the unit, since it is ‘TOSHIBA EXT’ and the part ‘TOSHIBA+space’ gets lost eventually
sct_image: error: unrecognized arguments: EXT/T1.nii
Can you confirm your results of sct_propseg?
When using sct_deepseg_sc, this does not happen. Could be possible to include the -CSF option to that module also?
Thank you very much!