i use the command of ‘sct_deepseg_gm ’for t2s,the results in the follow, i think the performance is not good
t2s.nii.gz (3.4 MB)
t2s_gmseg.nii.gz (34.0 KB)
Note: My deepest apologies for the late response. (The forum’s email notification system has been down, and so we missed your post. The notification system has since been fixed, and we are catching up on missed replies.)
Hi @Xujian_Wang,
I agree that the performance of sct_deepseg_gm appears to be quite poor on your data:
From what I can tell, though, the contrast between the GM and WM is quite low in your data:
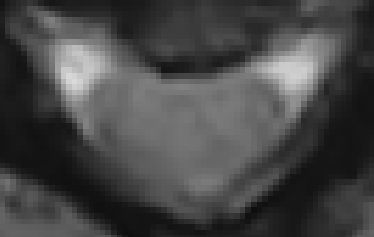
Comparing this to a sample T2s image in SCT’s testing dataset, which has a much higher contrast between the GM and WM regions.
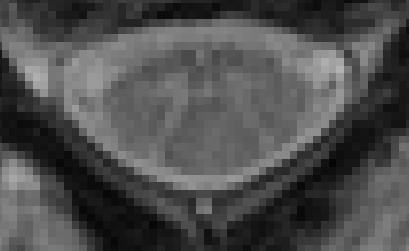
So, this may be a case where the performance of SCT’s gray matter segmentation algorithm is limited by the quality of the input data.
However, I’ll tag @jcohenadad, as he may have further input on how to improve the quality of the segmentation.
Kind regards,
Joshua
Hi! Sorry I’m late to the party. I find the segmentation QC shown here are actually quite good considering the relatively low GM/WM contrast!
@Xujian_Wang Could you please elaborate on the low performance?
As you said, my result is not bad?
Hi @Xujian_Wang,
My apologies, but I don’t understand what you are asking here.
Are you agreeing that the results are good? Or are you disagreeing?
((Basically, I want to check if this issue is resolved, or if you have more questions that you need answered.))
Kind regards,
Joshua
sorry for my question, i want to know that,could my results be used to analysis for research about gm
Yes, in principle, they could. But of course the definite answer depends on a variety of factors, including the underlying research hypotheses, the type of pathology you are dealing with, the kind of statistical analysis you are planning to do, the expected effect size, group size, etc.