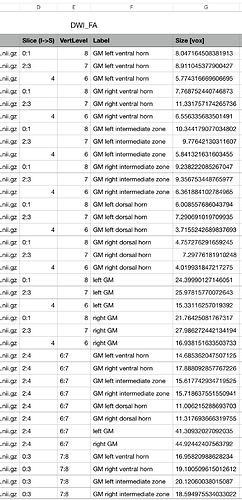
dear jcohenadad . I have a problem , when I run the outcome of the same dti . different I-S value shows the different outcome , even they are the same verlevel. what is the meaning of I-S in the csa document ? why it will come out?
at the same time , I am afraid I have problem in label c2,c2-c3 , c3 ,c3-c4. this is my script. could you have a look ?
!/usr/bin/env bash
file_dwi="TANG_WING_KUN_WIP_DTI_thk7_breath_1_5_2"
file_bvec=${file_dwi}.bvec
file_bval=${file_dwi}.bval
Separate b=0 and DW images
sct_dmri_separate_b0_and_dwi -i ${file_dwi}.nii.gz -bvec ${file_bvec}
sct_propseg -i ${file_dwi}_dwi_mean.nii.gz -c dwi
sct_create_mask -i ${file_dwi}_dwi_mean.nii.gz -p centerline,${file_dwi}_dwi_mean_seg.nii.gz -size 30mm
sct_crop_image -i ${file_dwi}.nii.gz -m mask_${file_dwi}_dwi_mean.nii.gz -o ${file_dwi}_crop.nii.gz
sct_dmri_moco -i ${file_dwi}_crop.nii.gz -bvec ${file_dwi}.bvec -x spline
file_dwi=${file_dwi}_crop_moco
file_dwi_mean=${file_dwi}_dwi_mean
# # Segment spinal cord (only if it does not exist)
sct_deepseg_sc -i ${file_dwi_mean}.nii.gz -c dwi -qc qc
file_dwi_seg=${file_dwi_mean}_seg
# # # Create labels, assuming that the mid-FOV is centered at C2 vertebral level
# sct_label_utils -i ${file_dwi_seg}.nii.gz -create-seg 2,3 -o label_C2.nii.gz
sct_label_utils -i ${file_dwi_seg}.nii.gz -create-seg -2,3 -o label_C2.nii.gz
sct_register_to_template -i ${file_dwi_mean}.nii.gz -s ${file_dwi_seg}.nii.gz -l label_C3.nii.gz -ref subject -c t1 -param step=1,type=seg,algo=centermassrot:step=2,type=im,algo=syn,metric=CC,slicewise=0,smooth=0,iter=3 -qc qc
mv warp_template2anat.nii.gz warp_template2dwi.nii.gz
mv warp_anat2template.nii.gz warp_dwi2template.nii.gz
sct_warp_template -d ${file_dwi_mean}.nii.gz -w warp_template2dwi.nii.gz -qc qc
sct_dmri_compute_dti -i ${file_dwi}.nii.gz -bvec ${file_bvec} -bval ${file_bval} -method standard
# sct_maths -i label/atlas/PAM50_atlas_00.nii.gz -add $(for i in `seq -w 2 2 36`; do echo "label/atlas/PAM50_atlas_${i}.nii.gz"; done) -o label/atlas/PAM50_atlas_left.nii.gz
# sct_maths -i label/atlas/PAM50_atlas_01.nii.gz -add $(for i in `seq -w 3 2 35`; do echo "label/atlas/PAM50_atlas_${i}.nii.gz"; done) -o label/atlas/PAM50_atlas_right.nii.gz
sct_extract_metric -i dti_FA.nii.gz -f label/atlas -l 30,31,32,33,34,35,58,59 -vert 2:3 -perlevel 1 -o DWI_FA.csv -append 1
sct_extract_metric -i dti_MD.nii.gz -f label/atlas -l 30,31,32,33,34,35,58,59 -vert 2:3 -perlevel 1 -o DWI_MD.csv -append 1
sct_extract_metric -i dti_RD.nii.gz -f label/atlas -l 30,31,32,33,34,35,58,59 -vert 2:3 -perlevel 1 -o DWI_RD.csv -append 1
sct_extract_metric -i dti_FA.nii.gz -f label/atlas -l 30,31,32,33,34,35,58,59 -vert 2:3 -o DWI_FA.csv -append 1
sct_extract_metric -i dti_FA.nii.gz -f label/atlas -l 30,31,32,33,34,35,58,59 -vert 3:4 -o DWI_FA.csv -append 1
sct_extract_metric -i dti_MD.nii.gz -f label/atlas -l 30,31,32,33,34,35,58,59 -vert 2:3 -o DWI_MD.csv -append 1
sct_extract_metric -i dti_MD.nii.gz -f label/atlas -l 30,31,32,33,34,35,58,59 -vert 3:4 -o DWI_MD.csv -append 1
sct_extract_metric -i dti_RD.nii.gz -f label/atlas -l 30,31,32,33,34,35,58,59 -vert 2:3 -o DWI_RD.csv -append 1
sct_extract_metric -i dti_RD.nii.gz -f label/atlas -l 30,31,32,33,34,35,58,59 -vert 3:4 -o DWI_RD.csv -append 1