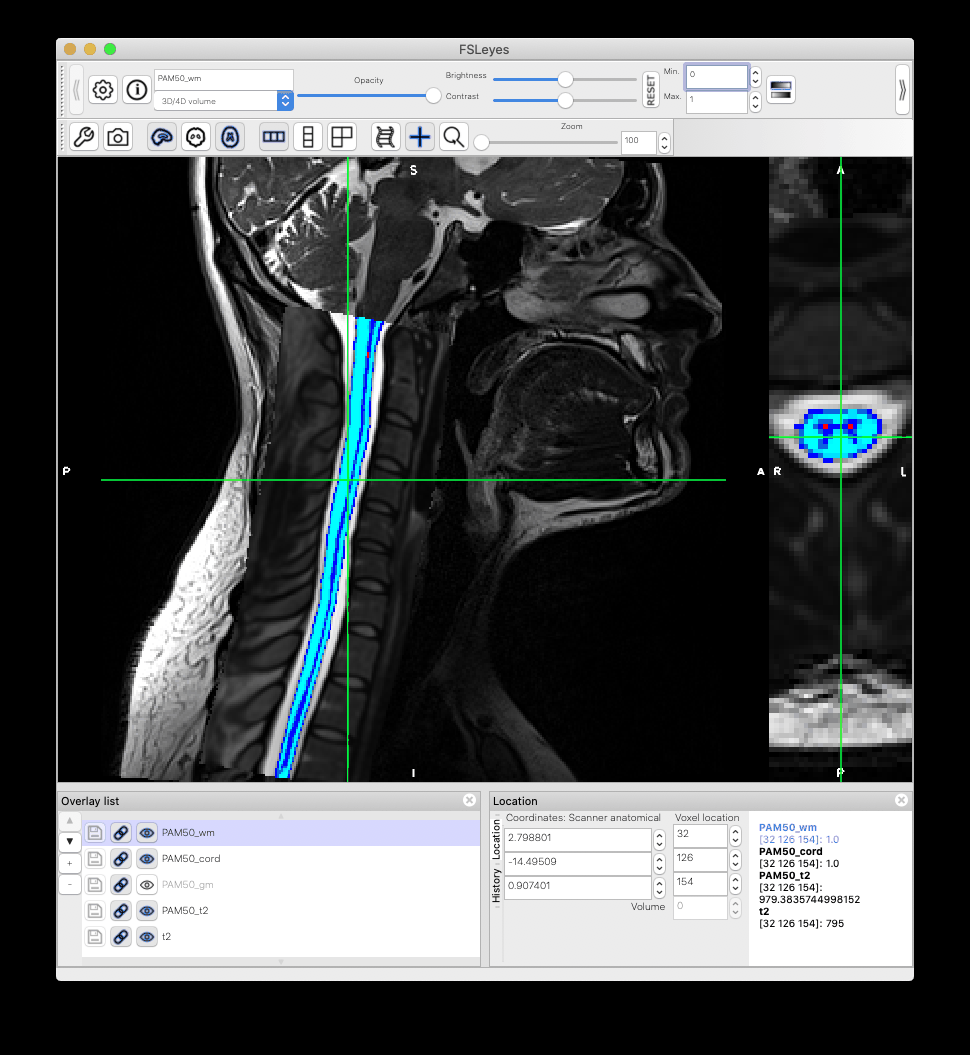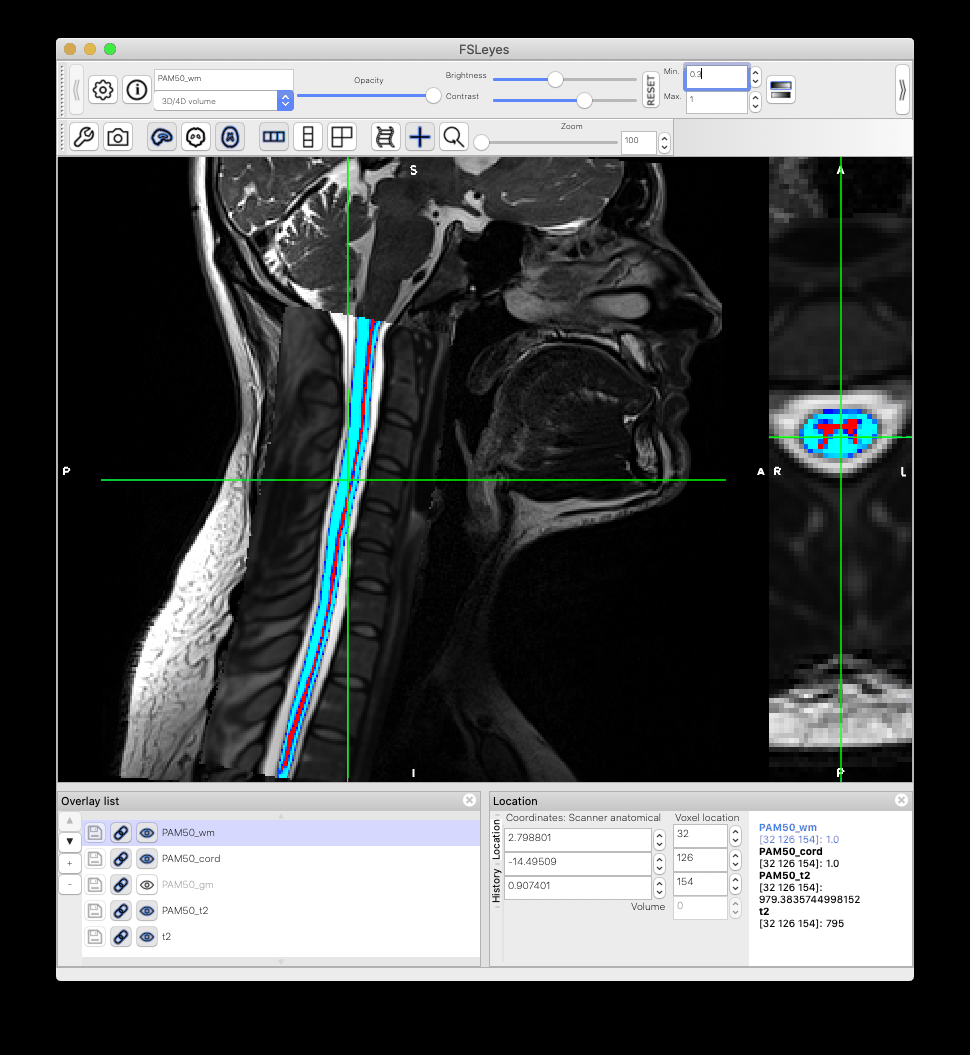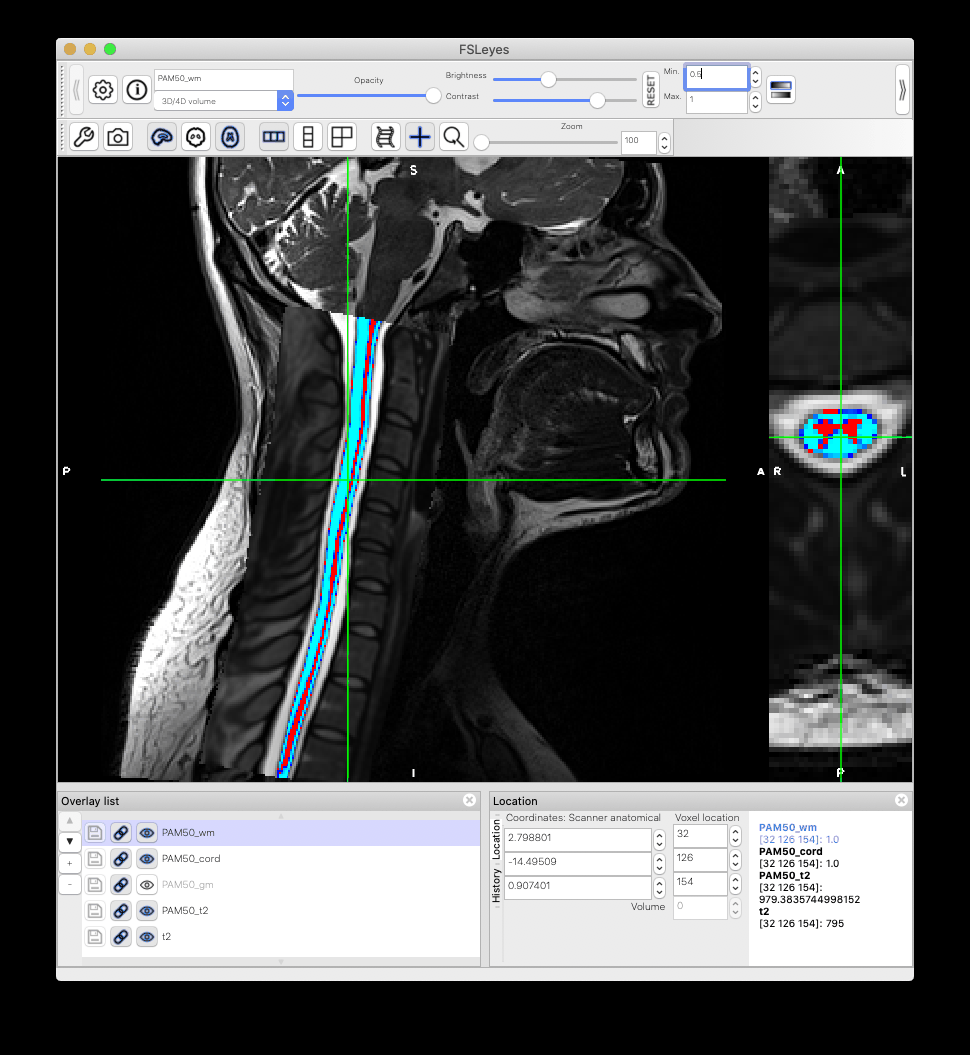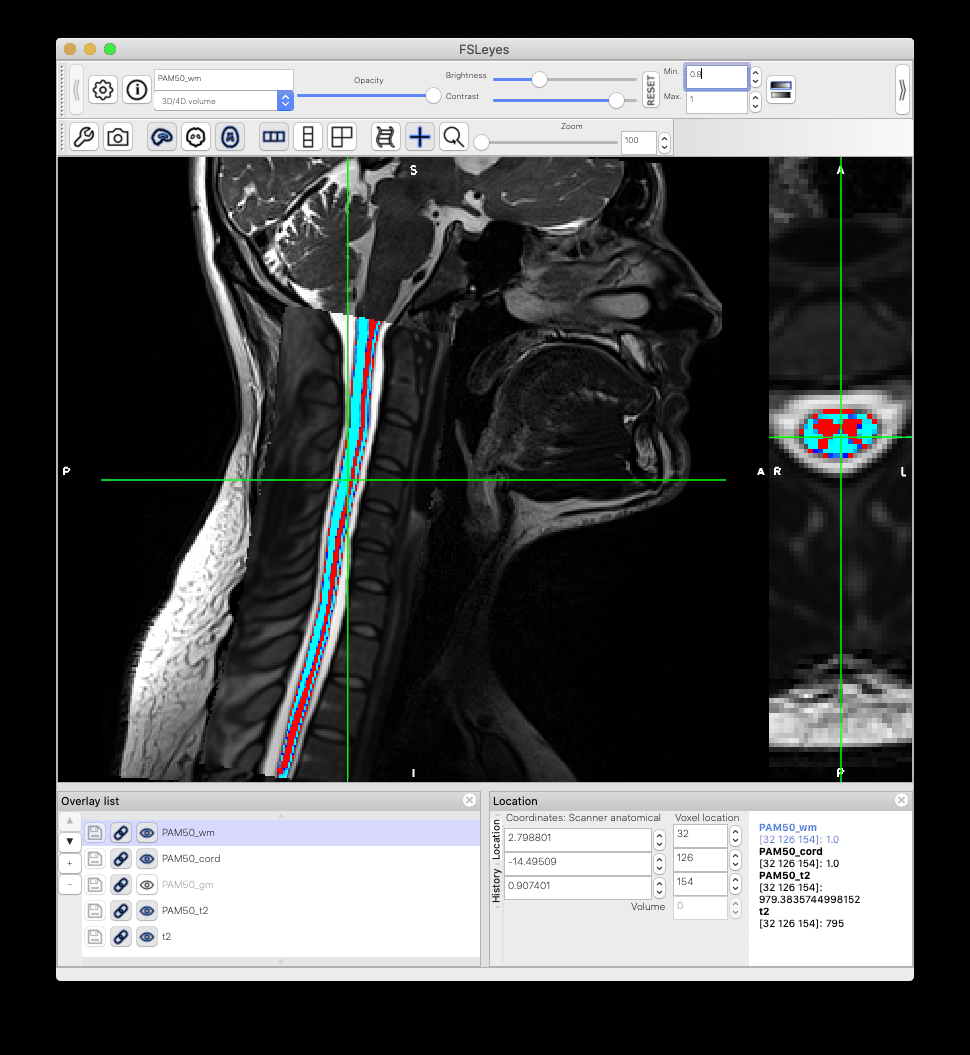
Dear experts,
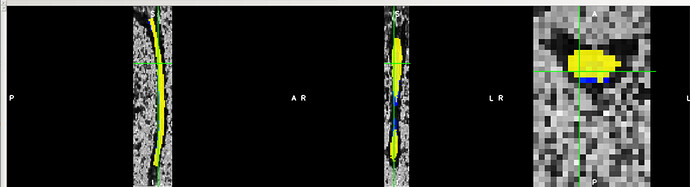
When I warp the template and all atlases to a native space image using the sct_warp_template script, I found that the atlas and the template PAM50_cord do not match exactly. What could be the reason?
Scripts that I used:
sct_register_multimodal -i ../XX-T2_SAG_resize_seg_labeled.nii -d XX_eddy_md_crop.nii -x nn -identity 1
sct_label_utils -i XX-T2_SAG_resize_seg_labeled_reg.nii -vert-body 1,10 -o XX_dmri_labels.nii.gz
sct_register_to_template -i XX_eddy_md_crop.nii -l XX_dmri_labels.nii.gz -s /home/jnavas/SPINALCORD/spine_masks/masks_reg_crop_fa/XX_WholeSpine_Mask2_reg.nii.gz -c t1 -param step=1,type=seg,algo=centermass:step=2,type=seg,algo=bsplinesyn,metric=MeanSquares,smooth=1,iter=6
sct_warp_template -d XX_eddy_md_crop.nii -w warp_template2anat.nii.gz -a 1 -s 1 -ofolder label