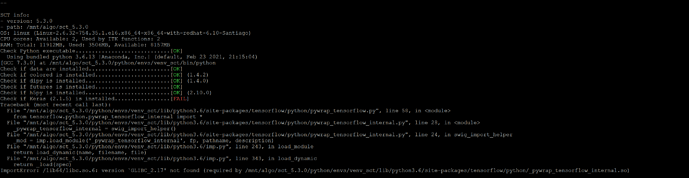
Also, here is ERROR log when trying to install SCT 3.2.3. Which is strange because we already had it installed, I just wanted to install it (AGAIN) as a sanity check before going hybrid. Good thing that I did because it doesnt seem to work now.
Building wheels for collected packages: nibabel, Keras, theano, unknown, html5lib, unknown
Running setup.py bdist_wheel for nibabel ... done
Stored in directory: /home/sradonjic/.cache/pip/wheels/a8/ba/6c/34dc8726562736c6f2ca41d324641035377832dc11d4b3168f
Running setup.py bdist_wheel for Keras ... done
Stored in directory: /home/sradonjic/.cache/pip/wheels/d1/70/83/9be5aef9c4c863ea21adacd0be83139b20d3d819401a2b07d3
Running setup.py bdist_wheel for theano ... done
Stored in directory: /home/sradonjic/.cache/pip/wheels/89/40/74/3a0b7d937890c66c4373120117ebf4ba99f4889b4a0a6a810c
Running setup.py bdist_wheel for unknown ... done
Stored in directory: /home/sradonjic/.cache/pip/wheels/9a/8e/50/89667f452c1817d655a618f551e5227f4ce65f882856bd6bc9
Running setup.py bdist_wheel for html5lib ... done
Stored in directory: /home/sradonjic/.cache/pip/wheels/50/ae/f9/d2b189788efcf61d1ee0e36045476735c838898eef1cad6e29
Running setup.py bdist_wheel for unknown ... error
Complete output from command /mnt/algo/sct_3.2.3_upgrade/python/bin/python -u -c "import setuptools, tokenize;__file__='/tmp/pip-build-BgWt8P/unknown/setup.py';f=getattr(tokenize, 'open', open)(__file__);code=f.read().replace('\r\n', '\n');f.close();exec(compile(code, __file__, 'exec'))" bdist_wheel -d /tmp/tmpDwMJiSpip-wheel- --python-tag cp27:
Traceback (most recent call last):
File "<string>", line 1, in <module>
IOError: [Errno 2] No such file or directory: '/tmp/pip-build-BgWt8P/unknown/setup.py'
----------------------------------------
Failed building wheel for unknown
Running setup.py clean for unknown
Complete output from command /mnt/algo/sct_3.2.3_upgrade/python/bin/python -u -c "import setuptools, tokenize;__file__='/tmp/pip-build-BgWt8P/unknown/setup.py';f=getattr(tokenize, 'open', open)(__file__);code=f.read().replace('\r\n', '\n');f.close();exec(compile(code, __file__, 'exec'))" clean --all:
Traceback (most recent call last):
File "<string>", line 1, in <module>
IOError: [Errno 2] No such file or directory: '/tmp/pip-build-BgWt8P/unknown/setup.py'
----------------------------------------
Failed cleaning build dir for unknown
Successfully built nibabel Keras theano unknown html5lib
Failed to build unknown
Installing collected packages: futures, nibabel, unknown, sympy, nipy, dipy, tqdm, contextlib2, raven, protobuf, backports.weakref, setuptools, markdown, werkzeug, html5lib, bleach, tensorflow-tensorboard, funcsigs, mock, tensorflow, theano, Keras, h5py
Found existing installation: setuptools 23.0.0
Uninstalling setuptools-23.0.0:
Exception:
Traceback (most recent call last):
File "/mnt/algo/sct_3.2.3_upgrade/python/lib/python2.7/site-packages/pip/basecommand.py", line 215, in main
status = self.run(options, args)
File "/mnt/algo/sct_3.2.3_upgrade/python/lib/python2.7/site-packages/pip/commands/install.py", line 342, in run
prefix=options.prefix_path,
File "/mnt/algo/sct_3.2.3_upgrade/python/lib/python2.7/site-packages/pip/req/req_set.py", line 778, in install
requirement.uninstall(auto_confirm=True)
File "/mnt/algo/sct_3.2.3_upgrade/python/lib/python2.7/site-packages/pip/req/req_install.py", line 754, in uninstall
paths_to_remove.remove(auto_confirm)
File "/mnt/algo/sct_3.2.3_upgrade/python/lib/python2.7/site-packages/pip/req/req_uninstall.py", line 115, in remove
renames(path, new_path)
File "/mnt/algo/sct_3.2.3_upgrade/python/lib/python2.7/site-packages/pip/utils/__init__.py", line 267, in renames
shutil.move(old, new)
File "/mnt/algo/sct_3.2.3_upgrade/python/lib/python2.7/shutil.py", line 302, in move
copy2(src, real_dst)
File "/mnt/algo/sct_3.2.3_upgrade/python/lib/python2.7/shutil.py", line 130, in copy2
copyfile(src, dst)
File "/mnt/algo/sct_3.2.3_upgrade/python/lib/python2.7/shutil.py", line 82, in copyfile
with open(src, 'rb') as fsrc:
IOError: [Errno 2] No such file or directory: '/mnt/algo/sct_3.2.3_upgrade/python/bin/easy_install-2.7'
You are using pip version 9.0.1, however version 21.1.2 is available.
You should consider upgrading via the 'pip install --upgrade pip' command.
pip install error with exit status 2. Check logs for more details
and verify your internet connection before reinstalling
Installation failed