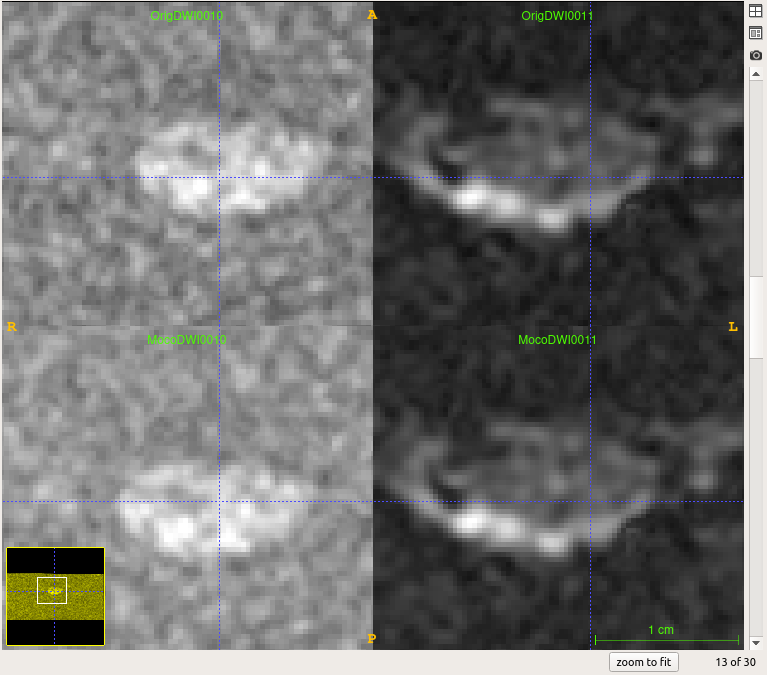
Dear SCT team,
I have dataset of dwi with several b-vals for IVIM analyses. There is a movement between first b-val and the last one (and of course between others too but lower).
Preproc:
fslsplit Ax_IVIM.nii.gz OrigDWI -t
fslmaths OrigDWI0008 -add OrigDWI0009 -add OrigDWI0010 -div 3 OrigDWI_mean
0008 b-val=300; 0009 = 400; 00010 = 800
sct_deepseg_sc -i OrigDWI_mean.nii.gz -c dwi -qc qc
sct_create_mask -i OrigDWI_mean.nii.gz -p centerline,OrigDWI_mean_seg.nii.gz -f cylinder -size 35mm
manual corr of mask if needed 0011=B0
itksnap -g OrigDWI_mean.nii.gz -s OrigDWI_mean_seg.nii.gz -o OrigDWI0011.nii.gz
I tried MoCo
sct_dmri_moco -i Ax_IVIM.nii.gz -m mask_DWI_mean.nii.gz -bvec Ax_IVIM.bvec -qc qc -qc-seg OrigDWI_mean_seg.nii.gz
but it does not help. So I found this problem (Sct_dmri_moco doesn't seem to correct motion) and tried p=0 but it did not help either
sct_dmri_moco -i Ax_IVIM.nii.gz -bvec Ax_IVIM.bvec -m mask_OrigDWI_mean.nii.gz -g 1 -param poly=0,metric=MI,smooth=1
Here are my dependencies:
ct_check_dependencies
–
Spinal Cord Toolbox (6.1)
sct_check_dependencies
SYSTEM INFORMATION
SCT info:
- version: 6.1
- path: /home/marek/sct_6.1
OS: linux (Linux-5.15.0-94-generic-x86_64-with-glibc2.31)
CPU cores: Available: 16, Used by ITK functions: 16
RAM: Total: 31980MB, Used: 2594MB, Available: 28882MB
OPTIONAL DEPENDENCIES
Check FSLeyes version…[OK] (1.3.0)
MANDATORY DEPENDENCIES
Check Python executable…[OK]
Using bundled python 3.9.18 (main, Sep 11 2023, 13:41:44)
[GCC 11.2.0] at /home/marek/sct_6.1/python/envs/venv_sct/bin/python
Check if data are installed…[OK]
Check if dipy is installed…[OK] (1.5.0)
Check if ivadomed is installed…[OK] (2.9.8)
Check if matplotlib is installed…[OK] (3.8.1)
Check if nibabel is installed…[OK] (3.2.2)
Check if nilearn is installed…[OK] (0.10.2)
Check if numpy is installed…[OK] (1.23.5)
Check if onnxruntime is installed…[OK] (1.16.1)
Check if pandas is installed…[OK] (1.4.4)
Check if portalocker is installed…[OK] (2.8.2)
Check if psutil is installed…[OK] (5.9.6)
Check if pyqt5 (5.12.3) is installed…[OK] (5.12.3)
Check if pytest is installed…[OK] (7.4.3)
Check if pytest-cov is installed…[OK] (4.1.0)
Check if requests is installed…[OK] (2.31.0)
Check if requirements-parser is installed…[OK]
Check if scipy is installed…[OK] (1.11.3)
Check if scikit-image is installed…[OK] (0.22.0)
Check if scikit-learn is installed…[OK] (1.3.2)
Check if xlwt is installed…[OK] (1.3.0)
Check if tqdm is installed…[OK] (4.66.1)
Check if transforms3d is installed…[OK] (0.4.1)
Check if urllib3 is installed…[OK] (2.0.7)
Check if pytest_console_scripts is installed…[OK]
Check if pyyaml is installed…[OK] (6.0.1)
Check if voxelmorph is installed…[OK] (0.2)
Check if wquantiles is installed…[OK] (0.4)
Check if xlsxwriter is installed…[OK] (3.1.9)
Check if spinalcordtoolbox is installed…[OK]
Check ANTs compatibility with OS …[OK]
Check PropSeg compatibility with OS …[OK]
Check if figure can be opened with PyQt…[OK]
Check if figure can be opened with matplotlib…[OK] (Using GUI backend: ‘QtAgg’)
Here are my testing data:
You can see that before MoCo (upper row 800 - 0) and after MoCo (lower row 800 - 0) there is some change in b800 image but still it is not registered with B0.
Please, can you help me?
Thank you,
Mark